5QJM
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z328695024 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QK2
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z54628578 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
4KMK
 
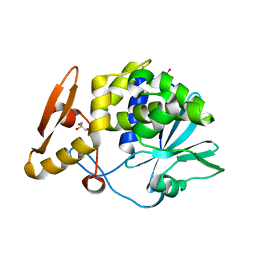 | | Crystal structure of Ribosome Inactivating protein from Momordica balsamina at 1.65 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, rRNA N-glycosidase | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Ribosome Inactivating protein from Momordica balsamina at 1.65 A resolution
To be Published
|
|
1FFJ
 
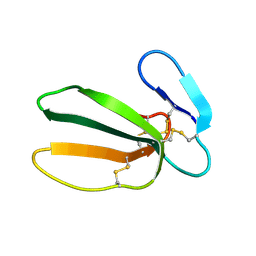 | | NMR STRUCTURE OF CARDIOTOXIN IN DPC-MICELLE | | Descriptor: | CYTOTOXIN 2 | | Authors: | Dubovskii, P.V, Dementieva, D.V, Bocharov, E.V, Utkin, Y.N, Arseniev, A.S. | | Deposit date: | 2000-07-25 | | Release date: | 2001-01-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Membrane binding motif of the P-type cardiotoxin.
J.Mol.Biol., 305, 2001
|
|
7LDE
 
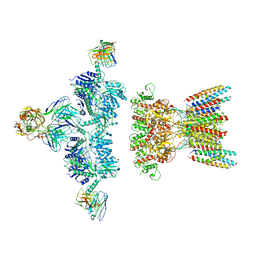 | | native AMPA receptor | | Descriptor: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | Authors: | Yu, J, Rao, P, Gouaux, E. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
7LDD
 
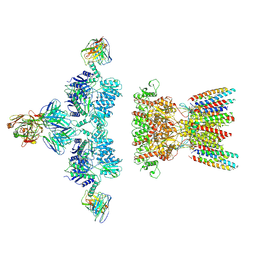 | | native AMPA receptor | | Descriptor: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | Authors: | Yu, J, Rao, P, Gouaux, E. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
1FEB
 
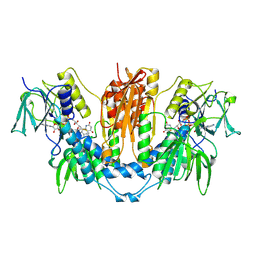 | |
7KC1
 
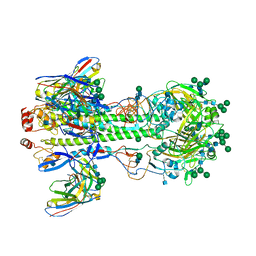 | |
4H2E
 
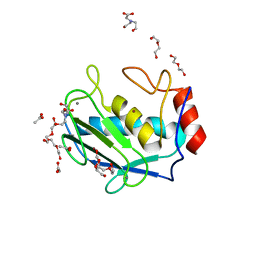 | | Crystal structure of an MMP twin inhibitor complexing two MMP-9 catalytic domains | | Descriptor: | ACETATE ION, BICINE, CALCIUM ION, ... | | Authors: | Stura, E.A, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Catalani, M.P, Dive, V, Rossello, A. | | Deposit date: | 2012-09-12 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
2HCA
 
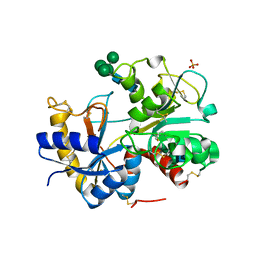 | | Crystal structure of bovine lactoferrin C-lobe liganded with Glucose at 2.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Prem Kumar, R, Ethayathulla, A.S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2006-06-15 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of bovine lactoferrin C-lobe liganded with Glucose at 2.8 A resolution
To be Published
|
|
1MX9
 
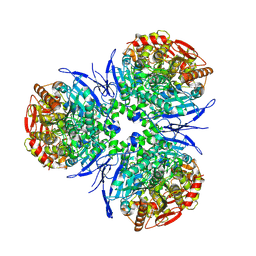 | | Crystal Structure of Human Liver Carboxylesterase in complexed with naloxone methiodide, a heroin analogue | | Descriptor: | (5A,17R)-4,5-EPOXY-3,14-DIHYDROXY-17-METHYL-6-OXO-17-(2-PROPENYL)-MORPHINANIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, liver Carboxylesterase I | | Authors: | Bencharit, S, Morton, C.L, Xue, Y, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2002-10-01 | | Release date: | 2003-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Heroin and Cocaine Metabolism by a Promiscuous Human Drug-Processing Enzyme
Nat.Struct.Biol., 10, 2003
|
|
3HZT
 
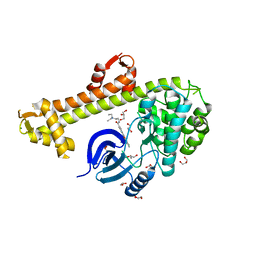 | | Crystal structure of Toxoplasma gondii CDPK3, TGME49_105860 | | Descriptor: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, Calcium-dependent protein kinase 3, GLYCEROL, ... | | Authors: | Wernimont, A.K, Artz, J.D, Finnerty, P, Wasney, G, Allali-Hassani, A, Vedadi, M, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-24 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1XJJ
 
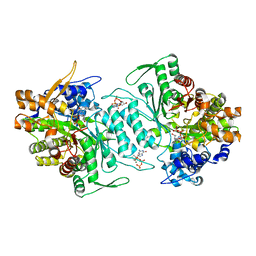 | | Structural mechanism of allosteric substrate specificity in a ribonucleotide reductase: dGTP complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ribonucleotide reductase, ... | | Authors: | Larsson, K.-M, Jordan, A, Eliasson, R, Reichard, P, Logan, D.T, Nordlund, P. | | Deposit date: | 2004-09-23 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Mechanism of Allosteric Substrate Specificity Regulation in a Ribonucleotide Reductase
Nat.Struct.Mol.Biol., 11, 2005
|
|
2HA6
 
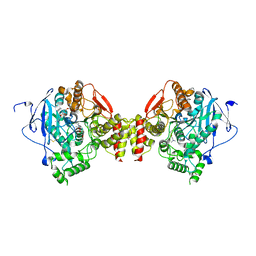 | | Crystal structure of mutant S203A of mouse acetylcholinesterase complexed with succinylcholine | | Descriptor: | 2,2'-[(1,4-DIOXOBUTANE-1,4-DIYL)BIS(OXY)]BIS(N,N,N-TRIMETHYLETHANAMINIUM), ACETIC ACID, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Radic, Z, Sulzenbacher, G, Kim, E, Taylor, P, Marchot, P. | | Deposit date: | 2006-06-12 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate and product trafficking through the active center gorge of acetylcholinesterase analyzed by crystallography and equilibrium binding
J.Biol.Chem., 281, 2006
|
|
2AWQ
 
 | |
7JV3
 
 | |
4BKO
 
 | | Enoyl-ACP reducatase FabV from Burkholderia pseudomallei (apo) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, PUTATIVE REDUCTASE BURPS305_1051, ... | | Authors: | Hirschbeck, M.W, Neckles, C, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-04-29 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Point Mutation Changes Substrate Binding Mechanism and Inhibitor Specificity of Yersinia Pestis Enoyl- Acp Reductase Fabv
To be Published
|
|
3HMJ
 
 | | Saccharomyces cerevisiae FAS type I | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Johansson, P, Mulinacci, B, Koestler, C, Vollrath, R, Oesterhelt, D, Grininger, M. | | Deposit date: | 2009-05-29 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Multimeric options for the auto-activation of the Saccharomyces cerevisiae FAS type I megasynthase
Structure, 17, 2009
|
|
1XJG
 
 | | Structural mechanism of allosteric substrate specificity in a ribonucleotide reductase: dATP-UDP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Larsson, K.-M, Jordan, A, Eliasson, R, Reichard, P, Logan, D.T, Nordlund, P. | | Deposit date: | 2004-09-23 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Mechanism of Allosteric Substrate Specificity Regulation in a Ribonucleotide Reductase
Nat.Struct.Mol.Biol., 11, 2004
|
|
7JZ1
 
 | |
4S0J
 
 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: 11BIF, 42F, 79S, and 123V mutant | | Descriptor: | Threonine--tRNA ligase | | Authors: | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2014-12-31 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
1RJ9
 
 | | Structure of the heterodimer of the conserved GTPase domains of the Signal Recognition Particle (Ffh) and Its Receptor (FtsY) | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Signal Recognition Protein, ... | | Authors: | Egea, P.F, Shan, S.O, Napetschnig, J, Savage, D.F, Walter, P, Stroud, R.M. | | Deposit date: | 2003-11-18 | | Release date: | 2004-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate twinning activates the signal recognition particle and its receptor
Nature, 427, 2004
|
|
1XPO
 
 | | Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic bicyclomycin | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*UP*CP*U)-3', BICYCLOMYCIN, MAGNESIUM ION, ... | | Authors: | Skordalakes, E, Brogan, A.P, Park, B.S, Kohn, H, Berger, J.M. | | Deposit date: | 2004-10-09 | | Release date: | 2005-02-08 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural mechanism of inhibition of the rho transcription termination factor by the antibiotic bicyclomycin
Structure, 13, 2005
|
|
2B8K
 
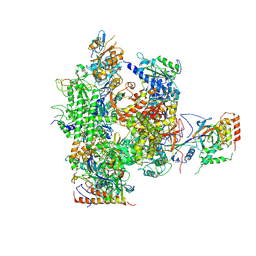 | | 12-subunit RNA Polymerase II | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, DNA-directed RNA polymerase II 19 kDa polypeptide, ... | | Authors: | Meyer, P.A, Ye, P, Zhang, M, Suh, M.H, Fu, J. | | Deposit date: | 2005-10-07 | | Release date: | 2006-06-20 | | Last modified: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Phasing RNA Polymerase II Using Intrinsically Bound Zn Atoms: An Updated Structural Model.
Structure, 14, 2006
|
|
4H35
 
 | |
