8D67
 
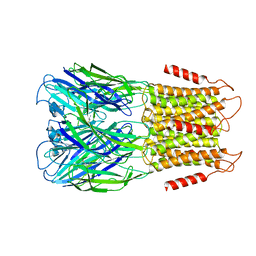 | | ELIC3 with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc | | 分子名称: | 2-AMINO-ETHANETHIOL, Erwinia ligand-gated ion channel | | 著者 | Petroff II, J.T, Deng, Z, Rau, M.J, Fitzpatrick, J.A.J, Yuan, P, Cheng, W.W.L. | | 登録日 | 2022-06-06 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Open-channel structure of a pentameric ligand-gated ion channel reveals a mechanism of leaflet-specific phospholipid modulation.
Nat Commun, 13, 2022
|
|
6NWD
 
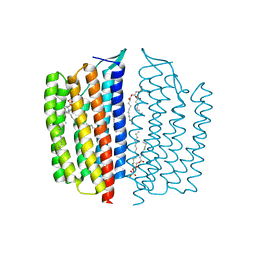 | | X-ray Crystallographic structure of Gloeobacter rhodopsin | | 分子名称: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DECANE, DODECANE, ... | | 著者 | Ernst, O.P, Morizumi, T, Ou, W.L. | | 登録日 | 2019-02-06 | | 公開日 | 2019-08-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray Crystallographic Structure and Oligomerization of Gloeobacter Rhodopsin.
Sci Rep, 9, 2019
|
|
4DDF
 
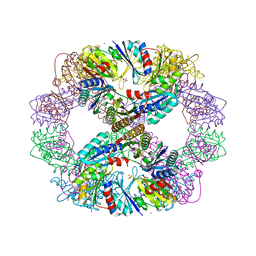 | | Computationally Designed Self-assembling Octahedral Cage protein, O333, Crystallized in space group P4 | | 分子名称: | CHLORIDE ION, Propanediol utilization polyhedral body protein PduT, SULFATE ION | | 著者 | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | 登録日 | 2012-01-18 | | 公開日 | 2012-06-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
4QOL
 
 | | Structure of Bacillus pumilus catalase | | 分子名称: | ACETATE ION, CHLORIDE ION, Catalase, ... | | 著者 | Loewen, P.C. | | 登録日 | 2014-06-20 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
6NWT
 
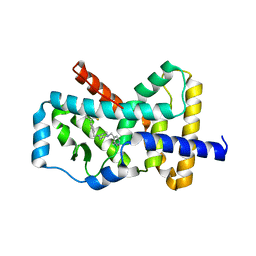 | | RORgamma Ligand Binding Domain | | 分子名称: | 1,1,1,3,3,3-hexafluoro-2-[2-fluoro-4'-({4-[(pyridin-4-yl)methyl]piperazin-1-yl}methyl)[1,1'-biphenyl]-4-yl]propan-2-ol, Nuclear receptor ROR-gamma | | 著者 | Strutzenberg, T.S, Park, H, Griffin, P.R. | | 登録日 | 2019-02-07 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
7X98
 
 | |
6VSP
 
 | | Structure of Serratia marcescens 2,3-butanediol dehydrogenase mutant Q247A | | 分子名称: | 1,2-ETHANEDIOL, 2,3-butanediol dehydrogenase, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Alahuhta, P.M, Lunin, V.V. | | 登録日 | 2020-02-11 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Phylogenetics-based identification and characterization of a superior 2,3-butanediol dehydrogenase for Zymomonas mobilis expression.
Biotechnol Biofuels, 13, 2020
|
|
6NXW
 
 | | Crystal structure of Arabidopsis thaliana cytosolic triosephosphate isomerase C218S mutant | | 分子名称: | Triosephosphate isomerase, cytosolic | | 著者 | Jimenez-Sandoval, P, Diaz-Quezada, C, Torres-Larios, A, Brieba, L.G. | | 登録日 | 2019-02-10 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for the modulation of plant cytosolic triosephosphate isomerase activity by mimicry of redox-based modifications.
Plant J., 99, 2019
|
|
7X3O
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07054 | | 分子名称: | (2~{R})-2-(3-fluoranyl-4-pyrimidin-5-yl-phenyl)butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | 登録日 | 2022-03-01 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07054
To Be Published
|
|
6HVL
 
 | | CdaA complex with c-di-AMP and AMP | | 分子名称: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ADENOSINE MONOPHOSPHATE, COBALT (II) ION, ... | | 著者 | Heidemann, J.L, Neumann, P, Ficner, R. | | 登録日 | 2018-10-11 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structures of the c-di-AMP-synthesizing enzyme CdaA.
J.Biol.Chem., 294, 2019
|
|
7X4P
 
 | | CD-NTase EfCdnE in complex with intermediate pppUpU | | 分子名称: | EfCdnE, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE, ... | | 著者 | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | 登録日 | 2022-03-03 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
5G4Q
 
 | | H.pylori Beta clamp in complex with 5-chloroisatin | | 分子名称: | 5-chloro-1H-indole-2,3-dione, DNA POLYMERASE III SUBUNIT BETA | | 著者 | Pandey, P, Gourinath, S. | | 登録日 | 2016-05-16 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Screening of E. coli beta-clamp Inhibitors Revealed that Few Inhibit Helicobacter pylori More Effectively: Structural and Functional Characterization.
Antibiotics (Basel), 7, 2018
|
|
6NWN
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with glutamic acid linked compound 10 | | 分子名称: | 5'-{[N-({(1S,2R)-2-[4-(carboxymethyl)benzene-1-carbonyl]cyclopentyl}acetyl)-L-gamma-glutamyl]amino}-2',5'-dideoxycytidine, ATP-dependent dethiobiotin synthetase BioD, CITRIC ACID | | 著者 | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | 登録日 | 2019-02-06 | | 公開日 | 2020-02-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.005 Å) | | 主引用文献 | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with glutamic acid linked compound 10
To Be Published
|
|
4LTA
 
 | |
4QNK
 
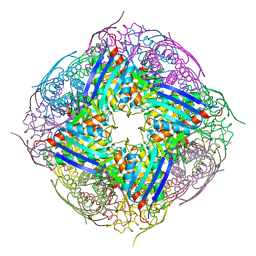 | | The structure of wt A. thaliana IGPD2 in complex with Mn2+ and phosphate | | 分子名称: | 1,2-ETHANEDIOL, Imidazoleglycerol-phosphate dehydratase 2, chloroplastic, ... | | 著者 | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R, Hawkes, T.R, Baker, P.J, Rice, D.W. | | 登録日 | 2014-06-18 | | 公開日 | 2015-06-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
4QOO
 
 | | Structure of Bacillus pumilus catalase with resorcinol bound. | | 分子名称: | CHLORIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Loewen, P.C. | | 登録日 | 2014-06-20 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
8D54
 
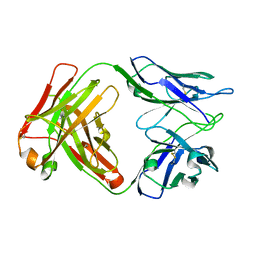 | |
4QOQ
 
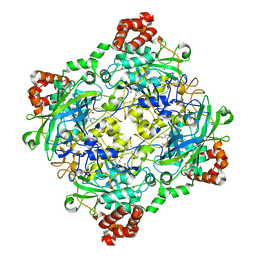 | | Structure of Bacillus pumilus catalase with guaiacol bound | | 分子名称: | CHLORIDE ION, Catalase, Guaiacol, ... | | 著者 | Loewen, P.C. | | 登録日 | 2014-06-20 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QON
 
 | | Structure of Bacillus pumilus catalase with catechol bound. | | 分子名称: | CATECHOL, CHLORIDE ION, Catalase, ... | | 著者 | Loewen, P.C. | | 登録日 | 2014-06-20 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
1GGK
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, ASN201HIS VARIANT. | | 分子名称: | CATALASE HPII, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | 登録日 | 2000-08-21 | | 公開日 | 2000-08-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
4QNS
 
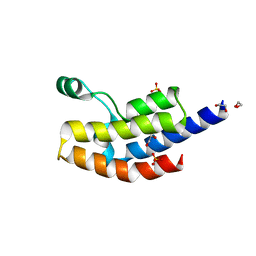 | | Crystal structure of bromodomain from Plasmodium faciparum GCN5, PF3D7_0823300 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Histone acetyltransferase GCN5, ... | | 著者 | Fonseca, M, Tallant, C, Knapp, S, Loppnau, P, von Delft, F, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Structural Genomics Consortium (SGC) | | 登録日 | 2014-06-18 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.497 Å) | | 主引用文献 | Crystal structure of bromodomain from Plasmodium faciparum GCN5, PF3D7_0823300
TO BE PUBLISHED
|
|
6W2A
 
 | | 1.65 A resolution structure of SARS-CoV 3CL protease in complex with inhibitor 7j | | 分子名称: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Replicase polyprotein 1a, [4,4-bis(fluoranyl)cyclohexyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | 著者 | Kashipathy, M.M, Lovell, S, Battaile, K.P, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-03-05 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
4QOR
 
 | | Structure of Bacillus pumilus catalase with chlorophenol bound. | | 分子名称: | 2-CHLOROPHENOL, CHLORIDE ION, Catalase, ... | | 著者 | Loewen, P.C. | | 登録日 | 2014-06-20 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
3DVZ
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. coli 23 S rRNA | | 分子名称: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | 著者 | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | 登録日 | 2008-07-21 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
6GI6
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with a Quinazolinone based ALK2 inhibitor with a 5-methyl core. | | 分子名称: | 1,2-ETHANEDIOL, 5-methyl-6-quinolin-5-yl-3~{H}-quinazolin-4-one, Activin receptor type-1, ... | | 著者 | Williams, E, Hudson, L, Bezerra, G.A, Sorrell, F, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | 登録日 | 2018-05-10 | | 公開日 | 2018-05-23 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
