6MGJ
 
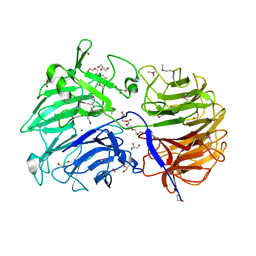 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Stogios, P.J, Skarina, T, Nocek, B, Arnal, G, Brumer, H, Savchenko, A. | | 登録日 | 2018-09-14 | | 公開日 | 2019-01-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
6AU1
 
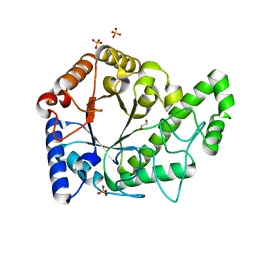 | | Structure of the PgaB (BpsB) glycoside hydrolase domain from Bordetella bronchiseptica | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative hemin storage protein, ... | | 著者 | Little, D.J, Bamford, N.C, Howell, P.L. | | 登録日 | 2017-08-30 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | PgaB orthologues contain a glycoside hydrolase domain that cleaves deacetylated poly-beta (1,6)-N-acetylglucosamine and can disrupt bacterial biofilms.
PLoS Pathog., 14, 2018
|
|
6YIF
 
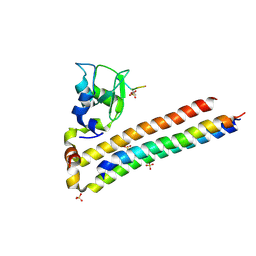 | | Structure of Chromosomal Passenger Complex (CPC) bound to phosphorylated Histone 3 peptide at 1.8 A. | | 分子名称: | Baculoviral IAP repeat-containing protein 5, Borealin, Inner centromere protein, ... | | 著者 | Serena, M, Elliott, P.R, Barr, F.A. | | 登録日 | 2020-04-01 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Molecular basis of MKLP2-dependent Aurora B transport from chromatin to the anaphase central spindle.
J.Cell Biol., 219, 2020
|
|
8Q4L
 
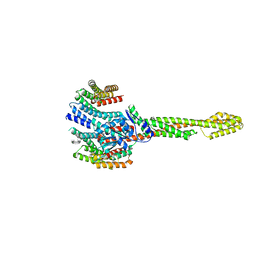 | | GBP1 bound by 14-3-3sigma | | 分子名称: | 14-3-3 protein sigma, Guanylate-binding protein 1 | | 著者 | Pfleiderer, M.M, Liu, X, Fisch, D, Anastasakou, E, Frickel, E.M, Galej, W.P. | | 登録日 | 2023-08-07 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (5.12 Å) | | 主引用文献 | PIM1 controls GBP1 activity to limit self-damage and to guard against pathogen infection.
Science, 382, 2023
|
|
6F38
 
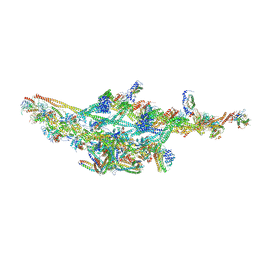 | | Cryo-EM structure of two dynein tail domains bound to dynactin and HOOK3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | 著者 | Lau, C.K, Urnavicius, L, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | 登録日 | 2017-11-28 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
5WNW
 
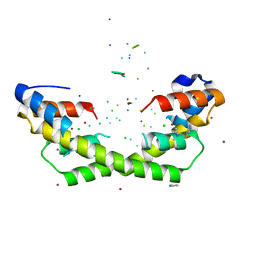 | | Chaperone Spy bound to Im7 6-45 ensemble | | 分子名称: | CHLORIDE ION, Colicin-E7 immunity protein, IMIDAZOLE, ... | | 著者 | Horowitz, S, Salmon, L, Koldewey, P, Ahlstrom, L.S, Martin, R, Xu, Q, Afonine, P.V, Trievel, R.C, Brooks, C.L, Bardwell, J.C.A. | | 登録日 | 2017-08-01 | | 公開日 | 2017-08-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
6JAO
 
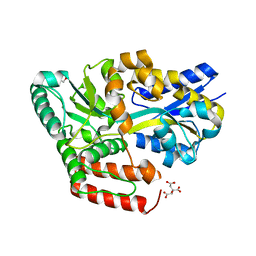 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein R356A in complex with palatinose | | 分子名称: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | 著者 | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | 登録日 | 2019-01-24 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
1BUS
 
 | |
6XZO
 
 | | Crystal structure of human carbonic anhydrase I in complex with 4-(3-(2-((4-bromo-2-hydroxybenzyl)amino)ethyl)ureido) benzenesulfonamide | | 分子名称: | 1-[2-[(4-bromanyl-2-oxidanyl-phenyl)methylamino]ethyl]-3-(4-sulfamoylphenyl)urea, Carbonic anhydrase 1, ZINC ION | | 著者 | Zanotti, G, Majid, A, Bozdag, M, Angeli, A, Carta, F, Berto, P, Supuran, C. | | 登録日 | 2020-02-05 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Benzylaminoethyureido-Tailed Benzenesulfonamides: Design, Synthesis, Kinetic and X-ray Investigations on Human Carbonic Anhydrases.
Int J Mol Sci, 21, 2020
|
|
3WRU
 
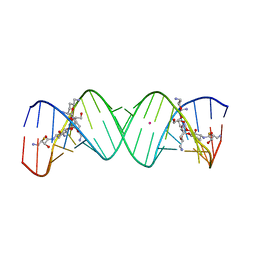 | | Crystal structure of the bacterial ribosomal decoding site in complex with synthetic aminoglycoside with F-HABA group | | 分子名称: | (2R,3R)-4-amino-N-[(1R,2S,3R,4R,5S)-5-amino-4-[(2,6-diamino-2,3,4,6-tetradeoxy-alpha-D-erythro-hexopyranosyl)oxy]-3-{[3-O-(2,6-diamino-2,3,4,6-tetradeoxy-beta-L-threo-hexopyranosyl)-beta-D-ribofuranosyl]oxy}-2-hydroxycyclohexyl]-3-fluoro-2-hydroxybutanamide, POTASSIUM ION, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | 著者 | Maianti, J.P, Kanazawa, H, Dozzo, P, Feeney, L.A, Armstrong, E.S, Kondo, J, Hanessian, S. | | 登録日 | 2014-02-27 | | 公開日 | 2014-11-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Toxicity Modulation, Resistance Enzyme Evasion, and A-Site X-ray Structure of Broad-Spectrum Antibacterial Neomycin Analogs
Acs Chem.Biol., 9, 2014
|
|
6B19
 
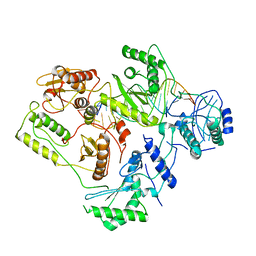 | | Architecture of HIV-1 reverse transcriptase initiation complex core | | 分子名称: | RNA genome fragment, reverse transcriptase p51 subunit, reverse transcriptase p66 subunit, ... | | 著者 | Larsen, K.P, Mathiharan, Y.K, Chen, D.H, Puglisi, J.D, Skiniotis, G, Puglisi, E.V. | | 登録日 | 2017-09-18 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Architecture of an HIV-1 reverse transcriptase initiation complex.
Nature, 557, 2018
|
|
7JY9
 
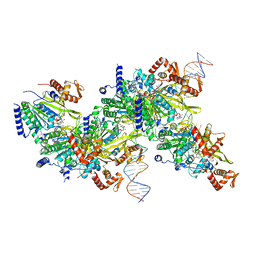 | |
5FES
 
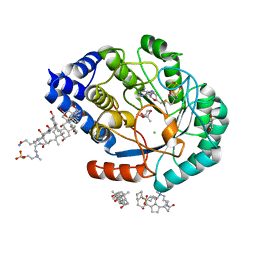 | | HydE from T. maritima in complex with (2R,4R)-MeSeTDA | | 分子名称: | (2~{R},4~{R})-2-methyl-1,3-selenazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | 著者 | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | 登録日 | 2015-12-17 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
5FF2
 
 | | HydE from T. maritima in complex with (2R,4R)-TDA | | 分子名称: | (2~{R},4~{R})-1,3-thiazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | 著者 | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | 登録日 | 2015-12-17 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
6Y3V
 
 | | 14-3-3 Sigma in complex with phosphorylated c-Jun peptide | | 分子名称: | 14-3-3 protein sigma, CALCIUM ION, MAGNESIUM ION, ... | | 著者 | Ballone, A, Lau, R.A, Zweipfenning, F.P.A, Ottmann, C. | | 登録日 | 2020-02-19 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.499 Å) | | 主引用文献 | A new soaking procedure for X-ray crystallographic structural determination of protein-peptide complexes.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
1E3K
 
 | | Human Progesteron Receptor Ligand Binding Domain in complex with the ligand metribolone (R1881) | | 分子名称: | (17BETA)-17-HYDROXY-17-METHYLESTRA-4,9,11-TRIEN-3-ONE, PROGESTERONE RECEPTOR | | 著者 | Matias, P.M, Donner, P, Coelho, R, Thomaz, M, Peixoto, C, Macedo, S, Otto, N, Joschko, S, Scholz, P, Wegg, A, Basler, S, Schafer, M, Egner, U, Carrondo, M.A. | | 登録日 | 2000-06-19 | | 公開日 | 2001-06-14 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations.
J. Biol. Chem., 275, 2000
|
|
6Y44
 
 | | 14-3-3 Sigma in complex with phosphorylated SOS1 peptide | | 分子名称: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Ballone, A, Lau, R.A, Zweipfenning, F.P.A, Ottmann, C. | | 登録日 | 2020-02-19 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | A new soaking procedure for X-ray crystallographic structural determination of protein-peptide complexes.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5WRN
 
 | |
7UNY
 
 | | Crystal structure of D2 nanobody in complex with PfCSS | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine-rich small secreted protein CSS, ... | | 著者 | Scally, S.W, Lim, P.S, Cowman, A.F. | | 登録日 | 2022-04-12 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (4.13 Å) | | 主引用文献 | PCRCR complex is essential for invasion of human erythrocytes by Plasmodium falciparum.
Nat Microbiol, 7, 2022
|
|
7TDZ
 
 | |
6JHR
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F6 | | 分子名称: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | 著者 | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | 登録日 | 2019-02-18 | | 公開日 | 2020-03-18 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
1DYR
 
 | | THE STRUCTURE OF PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE TO 1.9 ANGSTROMS RESOLUTION | | 分子名称: | DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIMETHOPRIM | | 著者 | Champness, J.N, Achari, A, Ballantine, S.P, Bryant, P.K, Delves, C.J, Stammers, D.K. | | 登録日 | 1994-09-14 | | 公開日 | 1995-10-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | The structure of Pneumocystis carinii dihydrofolate reductase to 1.9 A resolution.
Structure, 2, 1994
|
|
1E4D
 
 | | Structure of OXA10 beta-lactamase at pH 8.3 | | 分子名称: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, SULFATE ION | | 著者 | Maveyraud, L, Golemi, D, Kotra, L.P, Tranier, S, Vakulenko, S, Mobashery, S, Samama, J.P. | | 登録日 | 2000-07-03 | | 公開日 | 2001-01-12 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Insights Into Class D Beta-Lactamases are Revealed by the Crystal Structure of the Oxa10 Enzyme from Pseudomonas Aeruginosa
Structure, 8, 2000
|
|
6MU7
 
 | |
5F81
 
 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Roessler, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | 登録日 | 2015-12-08 | | 公開日 | 2016-02-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
