6N1V
 
 | |
6MPG
 
 | | Cryo-EM structure at 3.2 A resolution of HIV-1 fusion peptide-directed antibody, A12V163-b.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A12V163-b.01 Heavy Chain, ... | | 著者 | Acharya, P, Kwong, P.D. | | 登録日 | 2018-10-06 | | 公開日 | 2019-07-24 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | 分子名称: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | 著者 | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-09-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6PB1
 
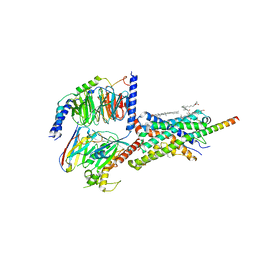 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 2 receptor in complex with Gs protein and Nb35 | | 分子名称: | CHOLESTEROL, Corticotropin-releasing factor receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | 登録日 | 2019-06-12 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-02-19 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
7QIW
 
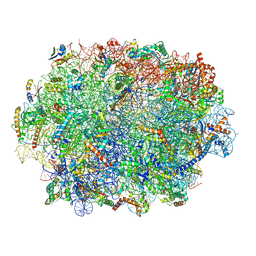 | | Specific features and methylation sites of a plant ribosome. 60S ribosomal subunit. | | 分子名称: | 25S rRNA, 5.8S rRNA, 50S ribosomal protein L22, ... | | 著者 | Cottilli, P, Itoh, Y, Amunts, A. | | 登録日 | 2021-12-16 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.35 Å) | | 主引用文献 | Cryo-EM structure and rRNA modification sites of a plant ribosome.
Plant Commun., 3, 2022
|
|
7QVP
 
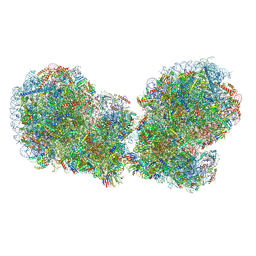 | | Human collided disome (di-ribosome) stalled on XBP1 mRNA | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Denk, T.G, Tesina, P, Beckmann, R. | | 登録日 | 2022-01-22 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | A distinct mammalian disome collision interface harbors K63-linked polyubiquitination of uS10 to trigger hRQT-mediated subunit dissociation.
Nat Commun, 13, 2022
|
|
6Q0C
 
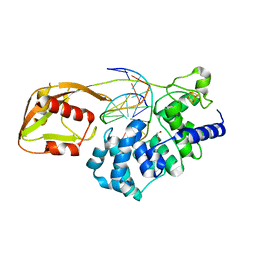 | | MutY adenine glycosylase bound to DNA containing a transition state analog (1N) paired with undamaged dG | | 分子名称: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, CALCIUM ION, ... | | 著者 | O'Shea Murray, V.L, Russelburg, L.P, Horvath, M.P, David, S.S. | | 登録日 | 2019-08-01 | | 公開日 | 2019-08-28 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for Finding OG Lesions and Avoiding Undamaged G by the DNA Glycosylase MutY.
Acs Chem.Biol., 15, 2020
|
|
6NF2
 
 | |
7OKX
 
 | | Structure of active transcription elongation complex Pol II-DSIF (SPT5-KOW5)-ELL2-EAF1 (composite structure) | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Chen, Y, Vos, S.M, Dienemann, C, Ninov, M, Urlaub, H, Cramer, P. | | 登録日 | 2021-05-18 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Allosteric transcription stimulation by RNA polymerase II super elongation complex.
Mol.Cell, 81, 2021
|
|
7ABP
 
 | |
8I23
 
 | | Clostridium thermocellum RNA polymerase transcription open complex with SigI1 and its promoter | | 分子名称: | DNA (80-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Li, J, Zhang, H, Li, D, Feng, Y, Zhu, P. | | 登録日 | 2023-01-13 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Structure of the transcription open complex of distinct sigma I factors.
Nat Commun, 14, 2023
|
|
8I24
 
 | | Clostridium thermocellum RNA polymerase transcription open complex with SigI6 and its promoter | | 分子名称: | DNA (80-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Li, J, Zhang, H, Li, D, Feng, Y, Zhu, P. | | 登録日 | 2023-01-13 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | Structure of the transcription open complex of distinct sigma I factors.
Nat Commun, 14, 2023
|
|
7OKY
 
 | | Structure of active transcription elongation complex Pol II-DSIF-ELL2-EAF1(composite structure) | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Chen, Y, Vos, S.M, Dienemann, C, Ninov, M, Urlaub, H, Cramer, P. | | 登録日 | 2021-05-18 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4.14 Å) | | 主引用文献 | Allosteric transcription stimulation by RNA polymerase II super elongation complex.
Mol.Cell, 81, 2021
|
|
7OL0
 
 | | Structure of active transcription elongation complex Pol II-DSIF (SPT5-KOW5) | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Chen, Y, Vos, S.M, Dienemann, C, Ninov, M, Urlaub, H, Cramer, P. | | 登録日 | 2021-05-18 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Allosteric transcription stimulation by RNA polymerase II super elongation complex.
Mol.Cell, 81, 2021
|
|
7OYG
 
 | | Dimeric form of SARS-CoV-2 RNA-dependent RNA polymerase | | 分子名称: | RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3'), RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3'), SARS-CoV-2 RNA-dependent RNA polymerase (nsp12), ... | | 著者 | Jochheim, F.A, Tegunov, D, Hillen, H.S, Schmitzova, J, Kokic, G, Dienemann, C, Cramer, P. | | 登録日 | 2021-06-24 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (5.5 Å) | | 主引用文献 | The structure of a dimeric form of SARS-CoV-2 polymerase
Communications Biology, 4, 2021
|
|
7QR9
 
 | | Crystal structure of CK1 delta in complex with PK-09-82 | | 分子名称: | 1,2-ETHANEDIOL, 4-[5-(4-fluorophenyl)-3-(pyridin-4-ylmethyl)imidazol-4-yl]-1~{H}-pyrrolo[2,3-b]pyridine, Casein kinase I isoform delta, ... | | 著者 | Chaikuad, A, Khirsariya, P, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2022-01-10 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery of Potent and Exquisitely Selective Inhibitors of Kinase CK1 with Tunable Isoform Selectivity.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7QRB
 
 | | Crystal structure of CK1 delta in complex with PK-09-129 | | 分子名称: | 3-(dimethylamino)-~{N}-[4-[4-(4-fluorophenyl)-5-(1~{H}-pyrrolo[2,3-b]pyridin-4-yl)imidazol-1-yl]cyclohexyl]propane-1-sulfonamide, Casein kinase I isoform delta, SULFATE ION | | 著者 | Chaikuad, A, Khirsariya, P, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2022-01-10 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Discovery of Potent and Exquisitely Selective Inhibitors of Kinase CK1 with Tunable Isoform Selectivity.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7R7S
 
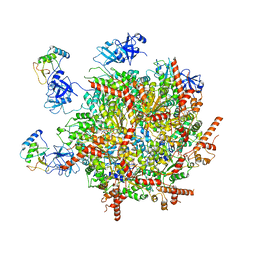 | | p47-bound p97-R155H mutant with ATPgammaS | | 分子名称: | NSFL1 cofactor p47, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | 著者 | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | 登録日 | 2021-06-25 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.23 Å) | | 主引用文献 | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7R7T
 
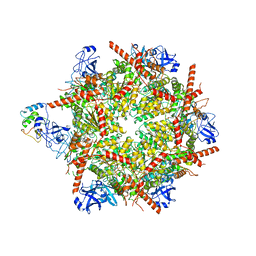 | | p47-bound p97-R155H mutant with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, NSFL1 cofactor p47, Transitional endoplasmic reticulum ATPase | | 著者 | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | 登録日 | 2021-06-25 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7OOP
 
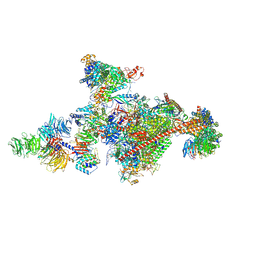 | | Pol II-CSB-CSA-DDB1-UVSSA-PAF-SPT6 (Structure 3) | | 分子名称: | DNA damage-binding protein 1, DNA excision repair protein ERCC-6, DNA excision repair protein ERCC-8, ... | | 著者 | Kokic, G, Cramer, P. | | 登録日 | 2021-05-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OO3
 
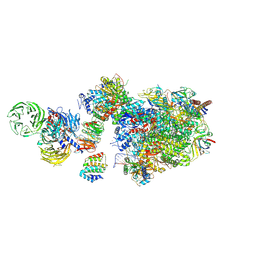 | | Pol II-CSB-CSA-DDB1-UVSSA (Structure1) | | 分子名称: | CSB element, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | 著者 | Kokic, G, Cramer, P. | | 登録日 | 2021-05-26 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OPD
 
 | |
7OOB
 
 | |
7OPC
 
 | |
8DVP
 
 | | Glycosylase MutY variant N146S in complex with DNA containing d(8-oxo-G) paired with substrate purine | | 分子名称: | ACETATE ION, Adenine DNA glycosylase, CALCIUM ION, ... | | 著者 | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | 登録日 | 2022-07-29 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
