3UVA
 
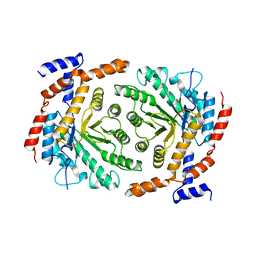 | | Crystal structure of L-rhamnose isomerase mutant W38F from Bacillus halodurans in complex with Mn | | 分子名称: | L-Rhamnose isomerase, MANGANESE (II) ION | | 著者 | Doan, T.T.N, Prabhu, P, Jeya, M, Kim, J.K, Kang, L.W, Lee, J.K. | | 登録日 | 2011-11-29 | | 公開日 | 2012-12-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
2AVJ
 
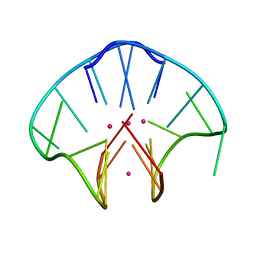 | | G4(Br)UTTG4 dimeric quadruplex | | 分子名称: | 5'-D(*GP*GP*GP*GP*(BRU)P*TP*TP*GP*GP*GP*G)-3', CACODYLATE ION, POTASSIUM ION | | 著者 | Hazel, P, Parkinson, G.N, Neidle, S. | | 登録日 | 2005-08-30 | | 公開日 | 2006-03-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Topology variation and loop structural homology in crystal and simulated structures of a bimolecular DNA quadruplex.
J.Am.Chem.Soc., 128, 2006
|
|
3UN2
 
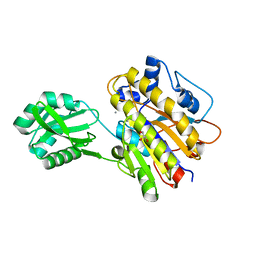 | | Phosphopentomutase T85Q variant enzyme | | 分子名称: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | 著者 | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | 登録日 | 2011-11-15 | | 公開日 | 2012-02-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
3UWN
 
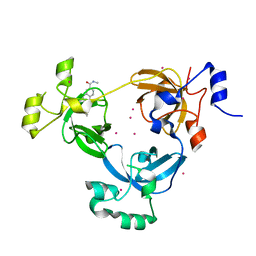 | | The 3-MBT repeat domain of L3MBTL1 in complex with a methyl-lysine mimic | | 分子名称: | Lethal(3)malignant brain tumor-like protein 1, UNKNOWN ATOM OR ION, [2-(phenylamino)benzene-1,4-diyl]bis{[4-(pyrrolidin-1-yl)piperidin-1-yl]methanone} | | 著者 | Zhong, N, Tempel, W, Wernimont, A.K, Graslund, S, Ingerman, L.A, Korboukh, V, Kireev, D.B, Gao, C, Frye, S.V, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | 登録日 | 2011-12-02 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The 3-MBT repeat domain of L3MBTL1 in complex with a methyl-lysine mimic
To be Published
|
|
3QKD
 
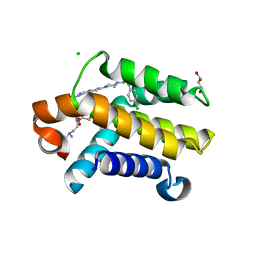 | | Crystal structure of Bcl-xL in complex with a Quinazoline sulfonamide inhibitor | | 分子名称: | (R)-N-(7-(4-((4'-chlorobiphenyl-2-yl)methyl)piperazin-1-yl)quinazolin-4-yl)-4-(4-(dimethylamino)-1-(phenylthio)butan-2-ylamino)-3-nitrobenzenesulfonamide, Bcl-2-like protein 1, CHLORIDE ION, ... | | 著者 | Czabotar, P.E, Smith, B.J. | | 登録日 | 2011-01-31 | | 公開日 | 2011-04-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Quinazoline sulfonamides as dual binders of the proteins B-cell lymphoma 2 and B-cell lymphoma extra long with potent proapoptotic cell-based activity.
J.Med.Chem., 54, 2011
|
|
8K37
 
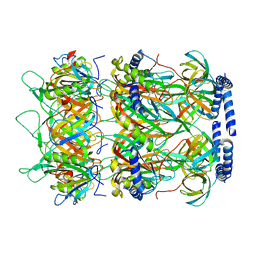 | | Structure of the bacteriophage lambda neck | | 分子名称: | Head-tail connector protein FII, Tail tube protein, Tail tube terminator protein | | 著者 | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | 登録日 | 2023-07-14 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
2UW6
 
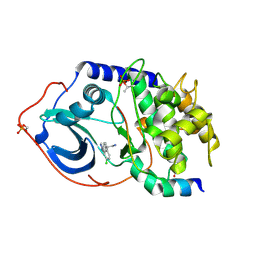 | | Structure of PKA-PKB chimera complexed with (S)-2-(4-chloro-phenyl)- 2-(4-1H-pyrazol-4-yl)-phenyl)-ethylamine | | 分子名称: | (2S)-2-(4-CHLOROPHENYL)-2-[4-(1H-PYRAZOL-4-YL)PHENYL]ETHANAMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | 著者 | Davies, T.G, Saxty, G, Woodhead, S.J, Berdini, V, Verdonk, M.L, Wyatt, P.G, Boyle, R.G, Barford, D, Downham, R, Garrett, M.D, Carr, R.A. | | 登録日 | 2007-03-19 | | 公開日 | 2007-05-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Identification of Inhibitors of Protein Kinase B Using Fragment-Based Lead Discovery
J.Med.Chem., 50, 2007
|
|
2O8K
 
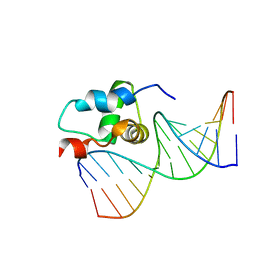 | | NMR Structure of the Sigma-54 RpoN Domain Bound to the-24 Promoter Element | | 分子名称: | 5'-D(*GP*AP*AP*AP*CP*GP*TP*GP*CP*CP*AP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*GP*CP*AP*CP*GP*TP*TP*TP*C)-3', RNA polymerase sigma factor RpoN | | 著者 | Doucleff, M, Pelton, J.G, Lee, P.S, Wemmer, D.E. | | 登録日 | 2006-12-12 | | 公開日 | 2007-07-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis of DNA recognition by the alternative sigma-factor, sigma54.
J.Mol.Biol., 369, 2007
|
|
3UP4
 
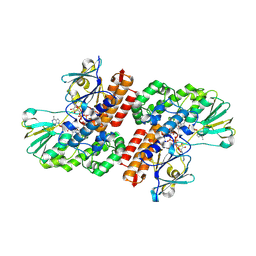 | | Crystal Structure of OTEMO complex with FAD and NADP (form 3) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | 著者 | Shi, R, Matte, A, Cygler, M, Lau, P. | | 登録日 | 2011-11-17 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.804 Å) | | 主引用文献 | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
8K39
 
 | | Structure of the bacteriophage lambda portal vertex | | 分子名称: | Major capsid protein, Portal protein B | | 著者 | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | 登録日 | 2023-07-14 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
5AHO
 
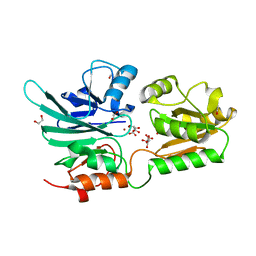 | | Crystal structure of human 5' exonuclease Apollo | | 分子名称: | 1,2-ETHANEDIOL, 5' EXONUCLEASE APOLLO, L(+)-TARTARIC ACID, ... | | 著者 | Allerston, C.K, Vollmar, M, Krojer, T, Pike, A.C.W, Newman, J.A, Carpenter, E, Quigley, A, Mahajan, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Gileadi, O. | | 登録日 | 2015-02-06 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | The Structures of the Snm1A and Snm1B/Apollo Nuclease Domains Reveal a Potential Basis for Their Distinct DNA Processing Activities.
Nucleic Acids Res., 43, 2015
|
|
5NJG
 
 | | Structure of an ABC transporter: part of the structure that could be built de novo | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3-Fab heavy chain, 5D3-Fab light chain, ... | | 著者 | Taylor, N.M.I, Manolaridis, I, Jackson, S.M, Kowal, J, Stahlberg, H, Locher, K.P. | | 登録日 | 2017-03-28 | | 公開日 | 2017-06-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | Structure of the human multidrug transporter ABCG2.
Nature, 546, 2017
|
|
2OAY
 
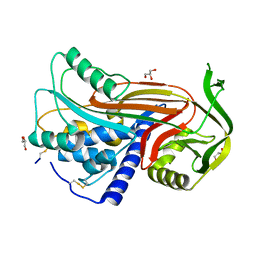 | | Crystal structure of latent human C1-inhibitor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Plasma protease C1 inhibitor | | 著者 | Harmat, V, Beinrohr, L, Gal, P, Dobo, J. | | 登録日 | 2006-12-18 | | 公開日 | 2007-05-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | C1 inhibitor serpin domain structure reveals the likely mechanism of heparin potentiation and conformational disease
J.Biol.Chem., 282, 2007
|
|
1JDK
 
 | | solution structure of lactam analogue (EDap) of HIV gp41 600-612 loop. | | 分子名称: | ACETYL GROUP | | 著者 | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | 登録日 | 2001-06-14 | | 公開日 | 2003-07-01 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
3NXQ
 
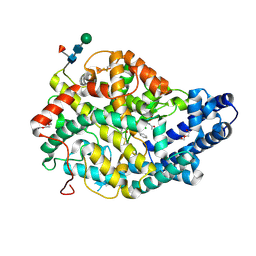 | | Angiotensin Converting Enzyme N domain glycsoylation mutant (Ndom389) in complex with RXP407 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, CHLORIDE ION, ... | | 著者 | Anthony, C.S, Corradi, H.R, Schwager, S.L.U, Redelinghuys, P, Georgiadis, D, Dive, V, Acharya, K.R, Sturrock, E.D. | | 登録日 | 2010-07-14 | | 公開日 | 2010-09-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | The N domain of human angiotensin-I-converting enzyme: the role of N-glycosylation and the crystal structure in complex with an N domain-specific phosphinic inhibitor, RXP407.
J.Biol.Chem., 285, 2010
|
|
7EXT
 
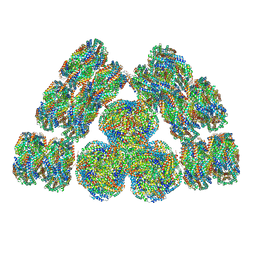 | | Cryo-EM structure of cyanobacterial phycobilisome from Synechococcus sp. PCC 7002 | | 分子名称: | Allophycocyanin alpha subunit, Allophycocyanin beta subunit, Allophycocyanin subunit alpha-B, ... | | 著者 | Zheng, L, Zheng, Z, Li, X, Wang, G, Zhang, K, Wei, P, Zhao, J, Gao, N. | | 登録日 | 2021-05-28 | | 公開日 | 2021-10-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural insight into the mechanism of energy transfer in cyanobacterial phycobilisomes.
Nat Commun, 12, 2021
|
|
2UW8
 
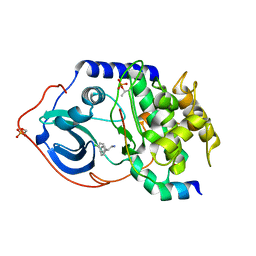 | | Structure of PKA-PKB chimera complexed with 2-(4-chloro-phenyl)-2- phenyl-ethylamine | | 分子名称: | (2R)-2-(4-CHLOROPHENYL)-2-PHENYLETHANAMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | 著者 | Davies, T.G, Saxty, G, Woodhead, S.J, Berdini, V, Verdonk, M.L, Wyatt, P.G, Boyle, R.G, Barford, D, Downham, R, Garrett, M.D, Carr, R.A. | | 登録日 | 2007-03-19 | | 公開日 | 2007-05-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Identification of Inhibitors of Protein Kinase B Using Fragment-Based Lead Discovery.
J.Med.Chem., 50, 2007
|
|
5AAD
 
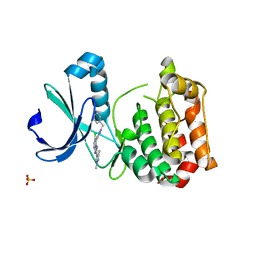 | | Aurora A kinase bound to an imidazopyridine inhibitor (7a) | | 分子名称: | 7-(1-benzyl-1H-pyrazol-4-yl)-6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridine, AURORA KINASE A, SULFATE ION | | 著者 | McIntyre, P.J, Kosmopoulou, M, Bayliss, R. | | 登録日 | 2015-07-24 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | 7-(Pyrazol-4-Yl)-3H-Imidazo[4,5-B]Pyridine-Based Derivatives for Kinase Inhibition: Co-Crystallisation Studies with Aurora-A Reveal Distinct Differences in the Orientation of the Pyrazole N1-Substituent.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7EYD
 
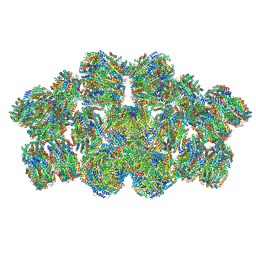 | | Cryo-EM structure of cyanobacterial phycobilisome from Anabaena sp. PCC 7120 | | 分子名称: | Allophycocyanin subunit alpha 1, Allophycocyanin subunit alpha-B, Allophycocyanin subunit beta, ... | | 著者 | Zheng, L, Zheng, Z, Li, X, Wang, G, Zhang, K, Wei, P, Zhao, J, Gao, N. | | 登録日 | 2021-05-30 | | 公開日 | 2021-10-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insight into the mechanism of energy transfer in cyanobacterial phycobilisomes.
Nat Commun, 12, 2021
|
|
2UW3
 
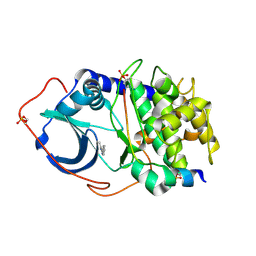 | | Structure of PKA-PKB chimera complexed with 5-methyl-4-phenyl-1H- pyrazole | | 分子名称: | 3-METHYL-4-PHENYL-1H-PYRAZOLE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | 著者 | Davies, T.G, Saxty, G, Woodhead, S.J, Berdini, V, Verdonk, M.L, Wyatt, P.G, Boyle, R.G, Barford, D, Downham, R, Garrett, M.D, Carr, R.A. | | 登録日 | 2007-03-19 | | 公開日 | 2007-05-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Identification of Inhibitors of Protein Kinase B Using Fragment-Based Lead Discovery
J.Med.Chem., 50, 2007
|
|
5NLM
 
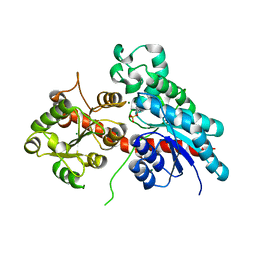 | | Complex between a UDP-glucosyltransferase from Polygonum tinctorium capable of glucosylating indoxyl and indoxyl sulfate | | 分子名称: | 3-SULFOOXY-1H-INDOLE, MAGNESIUM ION, indoxyl UDP-glucosyltransferase | | 著者 | Welner, D.H, Hsu, T, Dueber, J, Adams, P.D. | | 登録日 | 2017-04-04 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Employing a biochemical protecting group for a sustainable indigo dyeing strategy.
Nat. Chem. Biol., 14, 2018
|
|
2UUY
 
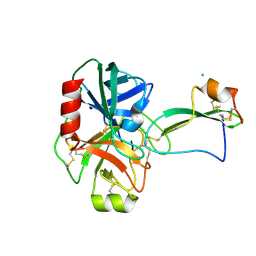 | | Structure of a tick tryptase inhibitor in complex with bovine trypsin | | 分子名称: | CALCIUM ION, CATIONIC TRYPSIN, CHLORIDE ION, ... | | 著者 | Siebold, C, Paesen, G.C, Harlos, K, Peacey, M.F, Nuttall, P.A, Stuart, D.I. | | 登録日 | 2007-03-08 | | 公開日 | 2007-04-10 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | A Tick Protein with a Modified Kunitz Fold Inhibits Human Tryptase.
J.Mol.Biol., 368, 2007
|
|
8K3N
 
 | |
4EVY
 
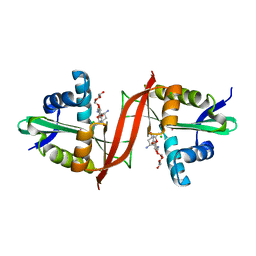 | | Crystal structure of aminoglycoside antibiotic 6'-N-acetyltransferase AAC(6')-Ig from Acinetobacter haemolyticus in complex with tobramycin | | 分子名称: | Aminoglycoside N(6')-acetyltransferase type 1, CHLORIDE ION, POTASSIUM ION, ... | | 著者 | Stogios, P.J, Evdokimova, E, Minasov, G, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-04-26 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.768 Å) | | 主引用文献 | Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes.
ACS Infect Dis., 3, 2017
|
|
5NBD
 
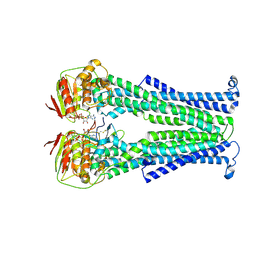 | | PglK flippase in complex with inhibitory nanobody | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Nanobody, WlaB protein | | 著者 | Perez, C, Pardon, E, Steyaert, J, Locher, K.P. | | 登録日 | 2017-03-01 | | 公開日 | 2017-05-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Structural basis of inhibition of lipid-linked oligosaccharide flippase PglK by a conformational nanobody.
Sci Rep, 7, 2017
|
|
