7MP2
 
 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH 1D-MYO-INOSITOL 1,5-BISDIPHOSPHATE TETRAKISPHOSPHATE (1,5-PP IP4) | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], S-arrestin | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2021-05-04 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7CSD
 
 | | AtPrR1 with NADP+ and (+)lariciresinol | | Descriptor: | 4-[[(3R,4R,5S)-4-(hydroxymethyl)-5-(3-methoxy-4-oxidanyl-phenyl)oxolan-3-yl]methyl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79709971 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
1JQ9
 
 | | Crystal structure of a complex formed between phospholipase A2 from Daboia russelli pulchella and a designed pentapeptide Phe-Leu-Ser-Tyr-Lys at 1.8 resolution | | Descriptor: | ACETIC ACID, Peptide inhibitor, Phospholipase A2 | | Authors: | Chandra, V, Jasti, J, Kaur, P, Dey, S, Betzel, C, Singh, T.P. | | Deposit date: | 2001-08-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Complex Formed between a Snake Venom Phospholipase A2 and a Potent Peptide Inhibitor Phe-Leu-Ser-Tyr-Lys at 1.8 A Resolution
J.BIOL.CHEM., 277, 2002
|
|
7MP0
 
 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH 1D-MYO-INOSITOL 5-DIPHOSPHATE PENTAKISPHOSPHATE (5-PP IP5) | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, 2-ETHOXYETHANOL, S-arrestin | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2021-05-04 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
5UJG
 
 | | ovGRN12-35_3s | | Descriptor: | Granulin | | Authors: | Bansal, P, Smout, M, Wilson, D, Caceres, C.C, Dastpeyman, M, Sotillo, J, Seifert, J, Brindley, P, Loukas, A, Daly, N. | | Deposit date: | 2017-01-17 | | Release date: | 2018-01-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Development of a Potent Wound Healing Agent Based on the Liver Fluke Granulin Structural Fold.
J. Med. Chem., 60, 2017
|
|
7CSF
 
 | | AtPrR1 with NADP+ and (-)secoisolariciresinol | | Descriptor: | (2R,3R)-2,3-bis[(3-methoxy-4-oxidanyl-phenyl)methyl]butane-1,4-diol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98241806 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
2P2I
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a nicotinamide inhibitor | | Descriptor: | N-(4-phenoxyphenyl)-2-[(pyridin-4-ylmethyl)amino]nicotinamide, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Kim, J.L, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2007-03-07 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evolution of a Highly Selective and Potent 2-(Pyridin-2-yl)-1,3,5-triazine Tie-2 Kinase Inhibitor
J.Med.Chem., 50, 2007
|
|
7MP1
 
 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH 1,5-DI-METHYLENEBISPHOSPHONATE INOSITOL TETRAKISPHOSPHATE (1,5-PCP-IP4) | | Descriptor: | S-arrestin, {[(1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl]bis[oxy(hydroxyphosphoryl)methanediyl]}bis(phosphonic acid) | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2021-05-04 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
3IHJ
 
 | | Human alanine aminotransferase 2 in complex with PLP | | Descriptor: | Alanine aminotransferase 2, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wisniewska, M, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Kotzsch, A, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human glutamate pyruvate transaminase 2
To be Published
|
|
7CSB
 
 | | AtPrR1 with NADP+ and (+)pinoresinol | | Descriptor: | 4-[(3S,3aR,6S,6aR)-6-(3-methoxy-4-oxidanyl-phenyl)-1,3,3a,4,6,6a-hexahydrofuro[3,4-c]furan-3-yl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.002017 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
5URN
 
 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and the transactivation domain 1 of p65 | | Descriptor: | RNA polymerase II transcription factor B subunit 1, Transcription factor p65 | | Authors: | Lecoq, L, Omichinski, J.G, Raiola, L, Cyr, N, Chabot, P, Arseneault, G, Legault, P. | | Deposit date: | 2017-02-11 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappa B and transcription regulatory factors.
Nucleic Acids Res., 45, 2017
|
|
4X9N
 
 | | Dithionite reduced L-alpha-Glycerophosphate Oxidase from Mycoplasma pneumoniae with FAD bound | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-alpha-glycerophosphate oxidase, NICKEL (II) ION | | Authors: | Elkhal, C.K, Kean, K.M, Parsonage, D, Claiborne, A, Karplus, P.A. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structure and proposed mechanism of l-alpha-glycerophosphate oxidase from Mycoplasma pneumoniae.
Febs J., 282, 2015
|
|
7LBF
 
 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Platelet-derived growth factor receptor alpha and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7BTU
 
 | | Crystal structure of TrmO from P. areuginosa | | Descriptor: | Putative tRNA (Adenine(37)-N6)-methyltransferase | | Authors: | Fan, C.P. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of TrmO
To Be Published
|
|
1SXN
 
 | | REDUCED BOVINE SUPEROXIDE DISMUTASE AT PH 5.0 | | Descriptor: | CALCIUM ION, COPPER (II) ION, CU, ... | | Authors: | Ferraroni, M, Rypniewski, W.R, Bruni, B, Orioli, P, Wilson, K.S, Mangani, S. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic determination of reduced bovine superoxide dismutase at pH 5.0 and of anion binding to its active site.
J.Biol.Inorg.Chem., 3, 1998
|
|
410D
 
 | | DUPLEX [5'-D(GCGTA+TACGC)]2 WITH INCORPORATED 2'-O-ETHOXYMETHYLENE RIBONUCLEOSIDE | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(T38)P*AP*CP*GP*C)-3'), SPERMINE | | Authors: | Tereshko, V, Portmann, S, Tay, E.C, Martin, P, Natt, F, Altmann, K.H, Egli, M. | | Deposit date: | 1998-06-30 | | Release date: | 1998-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Correlating structure and stability of DNA duplexes with incorporated 2'-O-modified RNA analogues.
Biochemistry, 37, 1998
|
|
412D
 
 | | DUPLEX [5'-D(GCGTA+TACGC)]2 WITH INCORPORATED 2'-O-METHYL-[TRI(OXYETHYL)] RIBONUCLEOSIDE | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(126)P*AP*CP*GP*C)-3'), MAGNESIUM ION | | Authors: | Tereshko, V, Portmann, S, Tay, E.C, Martin, P, Natt, F, Altmann, K.H, Egli, M. | | Deposit date: | 1998-06-30 | | Release date: | 1998-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Correlating structure and stability of DNA duplexes with incorporated 2'-O-modified RNA analogues.
Biochemistry, 37, 1998
|
|
5G2X
 
 | | Structure a of Group II Intron Complexed with its Reverse Transcriptase | | Descriptor: | 5'-R(*CP*AP*CP*AP*UP*CP*CP*AP*UP*AP*AP*CP)-3', GROUP II INTRON, GROUP II INTRON-ENCODED PROTEIN LTRA | | Authors: | Qu, G, Kaushal, P.S, Wang, J, Shigematsu, H, Piazza, C.L, Agrawal, R.K, Belfort, M, Wang, H.W. | | Deposit date: | 2016-04-16 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a Group II Intron in Complex with its Reverse Transcriptase.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7LWJ
 
 | |
4Y30
 
 | | Crystal structure of human protein arginine methyltransferase PRMT6 bound to SAH and EPZ020411 | | Descriptor: | GLYCEROL, MAGNESIUM ION, N,N'-dimethyl-N-({3-[4-({trans-3-[2-(tetrahydro-2H-pyran-4-yl)ethoxy]cyclobutyl}oxy)phenyl]-1H-pyrazol-4-yl}methyl)ethane-1,2-diamine, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aryl Pyrazoles as Potent Inhibitors of Arginine Methyltransferases: Identification of the First PRMT6 Tool Compound.
Acs Med.Chem.Lett., 6, 2015
|
|
6C0W
 
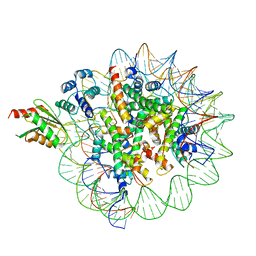 | | Cryo-EM structure of human kinetochore protein CENP-N with the centromeric nucleosome containing CENP-A | | Descriptor: | 147 mer DNA, Centromere protein N, Histone H2A, ... | | Authors: | Zhou, K, Pentakota, S, Vetter, I.R, Morgan, G.P, Petrovic, A, Musacchio, A, Luger, K. | | Deposit date: | 2018-01-02 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Decoding the centromeric nucleosome through CENP-N.
Elife, 6, 2017
|
|
7LWI
 
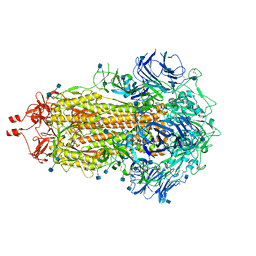 | |
7LWM
 
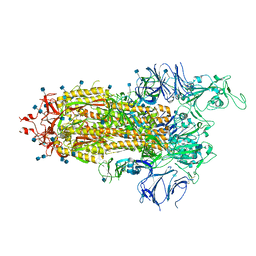 | |
2W2M
 
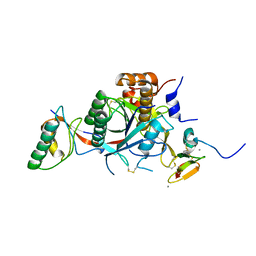 | | WT PCSK9-DELTAC BOUND TO WT EGF-A OF LDLR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 9 | | Authors: | Bottomley, M.J, Cirillo, A, Orsatti, L, Ruggeri, L, Fisher, T.S, Santoro, J.C, Cummings, R.T, Cubbon, R.M, Lo Surdo, P, Calzetta, A, Noto, A, Baysarowich, J, Mattu, M, Talamo, F, De Francesco, R, Sparrow, C.P, Sitlani, A, Carfi, A. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Biochemical Characterization of the Wild Type Pcsk9/Egf-Ab Complex and Natural Fh Mutants.
J.Biol.Chem., 284, 2009
|
|
7LWW
 
 | |
