6YEB
 
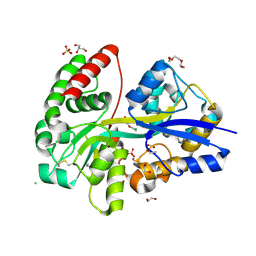 | | E.coli's Putrescine receptor PotF in its closed apo state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
8IBX
 
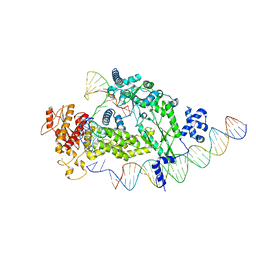 | | Structure of R2 with 3'UTR and DNA in unwinding state | | Descriptor: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBW
 
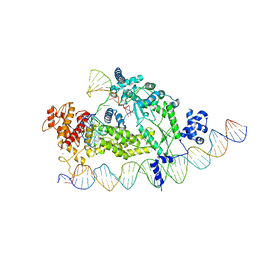 | | Structure of R2 with 3'UTR and DNA in binding state | | Descriptor: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
5KDT
 
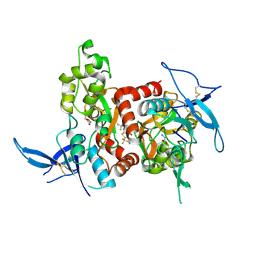 | | Structure of the human GluN1/GluN2A LBD in complex with GNE0723 | | Descriptor: | (1~{R},2~{R})-2-[7-[[5-chloranyl-3-(trifluoromethyl)pyrazol-1-yl]methyl]-5-oxidanylidene-2-(trifluoromethyl)-[1,3]thiazolo[3,2-a]pyrimidin-3-yl]cyclopropane-1-carbonitrile, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-06-08 | | Release date: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J. Med. Chem., 59, 2016
|
|
5GLS
 
 | | Structure of bovine Lactoperoxidase with a partially modified covalent bond with heme moiety | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tiwari, P, Singh, P.K, Sirohi, H.V, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-07-12 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of bovine lactoperoxidase with a partially linked heme moiety at 1.98 angstrom resolution
Biochim. Biophys. Acta, 1865, 2016
|
|
6YE7
 
 | | E.coli's Putrescine receptor PotF complexed with Cadaverine | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
6YGF
 
 | | NADase from Aspergillus fumigatus with trapped reaction products | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, AfNADase, ... | | Authors: | Stromland, O, Ziegler, M, Kallio, J.P. | | Deposit date: | 2020-03-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of fungal surface NADases predominantly present in pathogenic species.
Nat Commun, 12, 2021
|
|
4R48
 
 | | Racemic crystal structure of a calcium-bound DNA four-way junction | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3', CALCIUM ION, SODIUM ION | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Racemic DNA crystallography.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6MPX
 
 | | Twelve chloride ions induce formation and stabilize the NC1 hexamer of collagen IV assembled from transition state trimers | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bauer, R, Boudko, S.P, Hudson, B.G. | | Deposit date: | 2018-10-09 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A chloride ring is an ancient evolutionary innovation mediating the assembly of the collagen IV scaffold of basement membranes.
J.Biol.Chem., 294, 2019
|
|
5UEF
 
 | |
4R4P
 
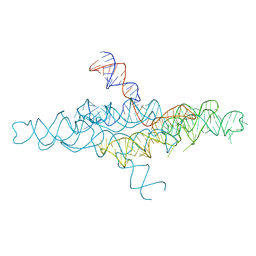 | | Crystal Structure of the VS ribozyme-A756G mutant | | Descriptor: | MAGNESIUM ION, VS ribozyme RNA | | Authors: | Piccirilli, J.A, Suslov, N.B, Dasgupta, S, Huang, H, Lilley, D.M.J, Rice, P.A. | | Deposit date: | 2014-08-19 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Crystal structure of the Varkud satellite ribozyme.
Nat.Chem.Biol., 11, 2015
|
|
5MQ0
 
 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | 3'-EXON OF UBC4 PRE-MRNA, BOUND BY PRP22 HELICASE, 5'-EXON OF UBC4 PRE-MRNA, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
6MS8
 
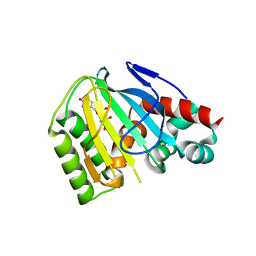 | | Crystal Structure of Chalcone Isomerase from Medicago Truncatula Complexed with (2S) Naringenin | | Descriptor: | Chalcone-flavonone isomerase family protein, NARINGENIN | | Authors: | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | Deposit date: | 2018-10-16 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
6MSO
 
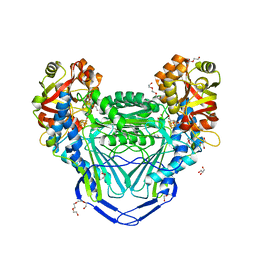 | | Crystal structure of mitochondrial fumarate hydratase from Leishmania major in a complex with inhibitor thiomalate | | Descriptor: | (2S)-2-sulfanylbutanedioic acid, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Feliciano, P.R, Drennan, C.L, Nonato, M.C. | | Deposit date: | 2018-10-17 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Crystal Structures of Fumarate Hydratases from Leishmania major in a Complex with Inhibitor 2-Thiomalate.
ACS Chem. Biol., 14, 2019
|
|
6N20
 
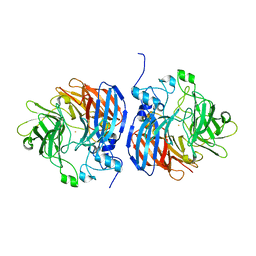 | | Structure of L509V CAO1 - growth condition 2 | | Descriptor: | CHLORIDE ION, Carotenoid oxygenase, FE (II) ION | | Authors: | Khadka, N, Shi, W, Kiser, P.D. | | Deposit date: | 2018-11-12 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evidence for distinct rate-limiting steps in the cleavage of alkenes by carotenoid cleavage dioxygenases.
J.Biol.Chem., 294, 2019
|
|
6Y4L
 
 | |
5MPS
 
 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-18 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
8EFN
 
 | | Structure of Sp-STING3 from Stylophora pistillata coral in complex with 3',3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Stimulator of interferon genes protein | | Authors: | Li, Y, Slavik, K.M, Morehouse, B.R, Mears, K, Kranzusch, P.J. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8EFM
 
 | | Structure of coral STING receptor from Stylophora pistillata in complex with 2',3'-cGAMP | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | Authors: | Li, Y, Slavik, K.M, Morehouse, B.R, Mears, K, Kranzusch, P.J. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
6CDE
 
 | | Cryo-EM structure at 3.8 A resolution of vaccine-elicited antibody vFP20.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, ... | | Authors: | Acharya, P, Xu, K, Liu, K, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
4QX2
 
 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6YDK
 
 | | Substrate-free P146A variant of beta-phosphoglucomutase from Lactococcus lactis | | Descriptor: | Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Wood, H.P, Cruz-Navarrete, F.A, Baxter, N.J, Trevitt, C.R, Robertson, A.J, Dix, S.R, Hounslow, A.M, Cliff, M.J, Waltho, J.P. | | Deposit date: | 2020-03-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Allomorphy as a mechanism of post-translational control of enzyme activity.
Nat Commun, 11, 2020
|
|
3CVL
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Phosphofructokinase (PFK) PTS1 peptide | | Descriptor: | Peroxisome targeting signal 1 receptor PEX5, T. brucei PFK PTS1 peptide Ac-HEELAKL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
4RCX
 
 | |
3CVQ
 
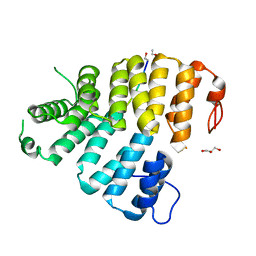 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to PTS1 peptide (7-SKL) | | Descriptor: | GLYCEROL, PTS1 peptide 7-SKL (Ac-SNRWSKL), Peroxisome targeting signal 1 receptor PEX5 | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
