7QQ5
 
 | |
7W43
 
 | |
2X5Z
 
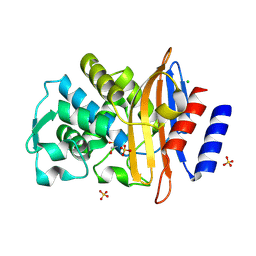
 | | Crystal structure of T. maritima GDP-mannose pyrophosphorylase in complex with GDP-mannose. | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, MANNOSE-1-PHOSPHATE GUANYLYLTRANSFERASE | | Authors: | Pelissier, M.C, Lesley, S, Kuhn, P, Bourne, Y. | | Deposit date: | 2010-02-12 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of Bacterial Guanosine-Diphospho-D-Mannose Pyrophosphorylase and its Regulation by Divalent Ions.
J.Biol.Chem., 285, 2010
|
|
7W46
 
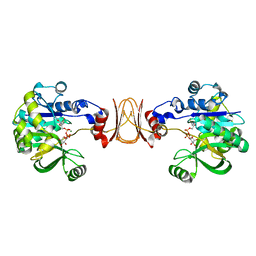
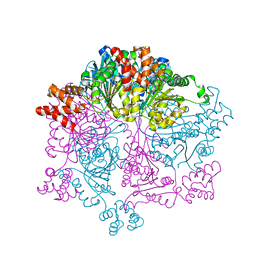 | | Crystal structure of Bacillus subtilis YjoB with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Uncharacterized ATPase YjoB | | Authors: | Dahal, P, Kwon, E, Kim, D.Y. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and biochemical analysis suggest that YjoB ATPase is a putative substrate-specific molecular chaperone.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1CFF
 
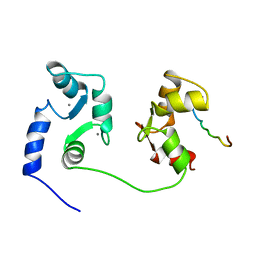 | | NMR SOLUTION STRUCTURE OF A COMPLEX OF CALMODULIN WITH A BINDING PEPTIDE OF THE CA2+-PUMP | | Descriptor: | CALCIUM ION, CALCIUM PUMP, CALMODULIN | | Authors: | Elshorst, B, Hennig, M, Foersterling, H, Diener, A, Maurer, M, Schulte, P, Schwalbe, H, Krebs, J, Schmid, H, Vorherr, T, Carafoli, E, Griesinger, C. | | Deposit date: | 1999-03-18 | | Release date: | 1999-09-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a complex of calmodulin with a binding peptide of the Ca2+ pump.
Biochemistry, 38, 1999
|
|
7Q6K
 
 | |
5YF8
 
 | |
7Q6J
 
 | | Crystal structure of the human GDAP1 CMT2 mutant-H123R | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ganglioside-induced differentiation-associated protein 1 | | Authors: | Sutinen, A, Kursula, P. | | Deposit date: | 2021-11-08 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into Charcot-Marie-Tooth disease-linked mutations in human GDAP1.
Febs Open Bio, 12, 2022
|
|
7W42
 
 | | Crystal structure of Bacillus subtilis YjoB | | Descriptor: | Uncharacterized ATPase YjoB | | Authors: | Dahal, P, Kwon, E, Kim, D.Y. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Crystal structure and biochemical analysis suggest that YjoB ATPase is a putative substrate-specific molecular chaperone.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W61
 
 | | Crystal structure of farnesol dehydrogenase from Helicoverpa armigera | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Farnesol dehydrogenase, ... | | Authors: | Kumar, R, Das, J, Mahto, J.K, Sharma, M, Kumar, P, Sharma, A.K. | | Deposit date: | 2021-12-01 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and molecular characterization of NADP + -farnesol dehydrogenase from cotton bollworm, Helicoverpaarmigera.
Insect Biochem.Mol.Biol., 147, 2022
|
|
3IM7
 
 | |
1CX9
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-AMINOPHENYLTHIO)-BUTYLPHOSPHONIC ACID | | Descriptor: | 4-(2-AMINOPHENYLTHIO)-BUTYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-08-29 | | Release date: | 1999-12-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
3IMO
 
 | | Structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS14 | | Descriptor: | ACETATE ION, Integron cassette protein | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2009-08-11 | | Release date: | 2009-08-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integron gene cassettes: a repository of novel protein folds with distinct interaction sites
Plos One, 8, 2013
|
|
7QWZ
 
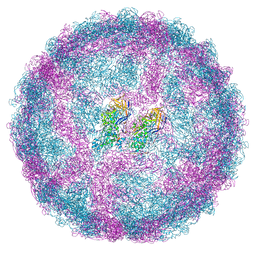 | | Full capsid of Saccharomyces cerevisiae virus L-BCLa | | Descriptor: | Major capsid protein | | Authors: | Grybchuk, D, Prochazkova, M, Fuzik, T, Konovalovas, A, Serva, S, Yurchenko, V, Plevka, P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of L-BC virus and its open particle provide insight into Totivirus capsid assembly.
Commun Biol, 5, 2022
|
|
7QVY
 
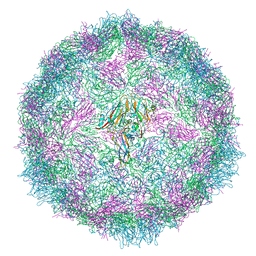 | | Cryo-EM structure of coxsackievirus A6 empty particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Buttner, C.R, Spurny, R, Fuzik, T, Plevka, P. | | Deposit date: | 2022-01-24 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Cryo-electron microscopy and image classification reveal the existence and structure of the coxsackievirus A6 virion.
Commun Biol, 5, 2022
|
|
7QVX
 
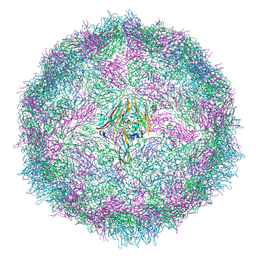 | | Cryo-EM structure of coxsackievirus A6 altered particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Buttner, C.R, Spurny, R, Fuzik, T, Plevka, P. | | Deposit date: | 2022-01-24 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-electron microscopy and image classification reveal the existence and structure of the coxsackievirus A6 virion.
Commun Biol, 5, 2022
|
|
1CZ2
 
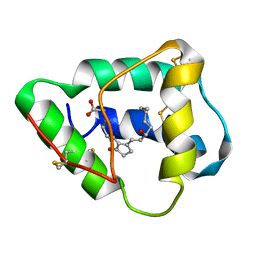 | | SOLUTION STRUCTURE OF WHEAT NS-LTP COMPLEXED WITH PROSTAGLANDIN B2. | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN, PROSTAGLANDIN B2 | | Authors: | Tassin-Moindrot, S, Caille, A, Douliez, J.-P, Marion, D, Vovelle, F. | | Deposit date: | 1999-09-01 | | Release date: | 2000-09-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The wide binding properties of a wheat nonspecific lipid transfer protein. Solution structure of a complex with prostaglandin B2.
Eur.J.Biochem., 267, 2000
|
|
7QWX
 
 | | Empty capsid of Saccharomyces cerevisiae virus L-BCLa | | Descriptor: | Major capsid protein | | Authors: | Grybchuk, D, Prochazkova, M, Fuzik, T, Konovalovas, A, Serva, S, Yurchenko, V, Plevka, P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of L-BC virus and its open particle provide insight into Totivirus capsid assembly.
Commun Biol, 5, 2022
|
|
7QW9
 
 | | Cryo-EM structure of coxsackievirus A6 mature virion | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Buttner, C.R, Spurny, R, Fuzik, T, Plevka, P. | | Deposit date: | 2022-01-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Cryo-electron microscopy and image classification reveal the existence and structure of the coxsackievirus A6 virion.
Commun Biol, 5, 2022
|
|
8AAB
 
 | | S148F mutant of blue-to-red fluorescent timer mRubyFT | | Descriptor: | mRubyFT S148F mutant of blue-to-red fluorescent timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Dorovatovskii, P.V, Subach, O.M, Popov, V.O, Subach, F.V. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | mRubyFT/S147I, a mutant of blue-to-red fluorescent timer
Crystallography Reports, 2022
|
|
2WUG
 
 | | Crystal structure of S114A mutant of HsaD from Mycobacterium tuberculosis in complex with HOPDA | | Descriptor: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, ... | | Authors: | Lack, N.A, Yam, K.C, Lowe, E.D, Horsman, G.P, Owen, R.L, Sim, E, Eltis, L.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a C-C Hydrolase from Mycobacterium Tuberuclosis Involved in Cholesterol Metabolism.
J.Biol.Chem., 285, 2010
|
|
5VLN
 
 | | NMR structure of the N-domain of troponin C bound to switch region of troponin I | | Descriptor: | Troponin C, slow skeletal and cardiac muscles,Troponin I, cardiac muscle | | Authors: | Cai, F, Hwang, P.M, Sykes, B.D. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures reveal details of small molecule binding to cardiac troponin.
J. Mol. Cell. Cardiol., 101, 2016
|
|
7QS9
 
 | |
7QD0
 
 | | Structure of the orange carotenoid protein from Planktothrix agardhii binding echinenone in the C2 space group | | Descriptor: | ACETATE ION, ARGININE, GLYCEROL, ... | | Authors: | Andreeva, E.A, Hartmann, E, Schlichting, I, Colletier, J.-P. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function-dynamics relationships in the peculiar Planktothrix PCC7805 OCP1: Impact of his-tagging and carotenoid type.
Biochim Biophys Acta Bioenerg, 1863, 2022
|
|
5W90
 
 | | FEZ-1 metallo-beta-lactamase from Legionella gormanii modelled with unknown ligand | | Descriptor: | FEZ-1 protein, GLYCEROL, SULFATE ION, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Papamicael, C, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2017-06-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-beta-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril.
J. Mol. Biol., 325, 2003
|
|
