8I9F
 
 | | S-RBD (Omicron BA.2.75) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8I9G
 
 | | S-RBD (Omicron BF.7) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8I9B
 
 | | S-ECD (Omicron BA.2.75) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
1D7S
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH DCS | | Descriptor: | D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
6NXR
 
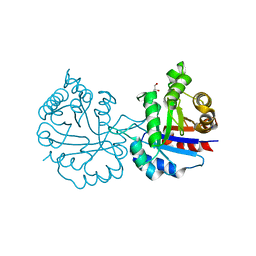 | | Crystal structure of Arabidopsis thaliana cytosolic triosephosphate isomerase C13D mutant | | Descriptor: | GLYCEROL, SODIUM ION, Triosephosphate isomerase, ... | | Authors: | Jimenez-Sandoval, P, Diaz-Quezada, C, Torres-Larios, A, Brieba, L.G. | | Deposit date: | 2019-02-09 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the modulation of plant cytosolic triosephosphate isomerase activity by mimicry of redox-based modifications.
Plant J., 99, 2019
|
|
6NXY
 
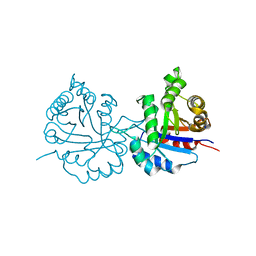 | | Crystal structure of Arabidopsis thaliana cytosolic triosephosphate isomerase C218D mutant | | Descriptor: | SODIUM ION, Triosephosphate isomerase, cytosolic | | Authors: | Jimenez-Sandoval, P, Diaz-Quezada, C, Torres-Larios, A, Brieba, L.G. | | Deposit date: | 2019-02-10 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural basis for the modulation of plant cytosolic triosephosphate isomerase activity by mimicry of redox-based modifications.
Plant J., 99, 2019
|
|
6O08
 
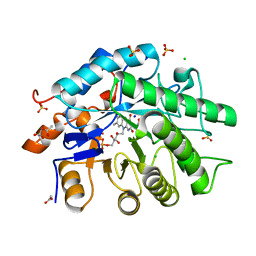 | | Gluconobacter Ene-Reductase (GluER) | | Descriptor: | ACETATE ION, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Garfinkle, S.E, Jeffrey, P, Hyster, T.K. | | Deposit date: | 2019-02-15 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Photoexcitation of flavoenzymes enables a stereoselective radical cyclization.
Science, 364, 2019
|
|
4ZPF
 
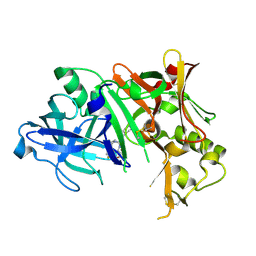 | |
4ZPG
 
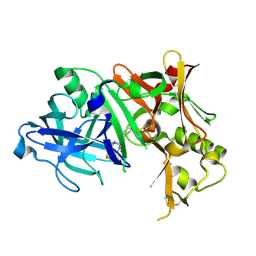 | |
1CII
 
 | |
1D7T
 
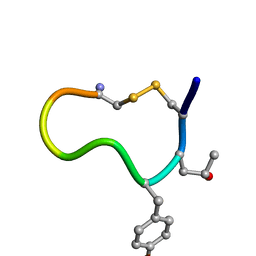 | |
1D8Y
 
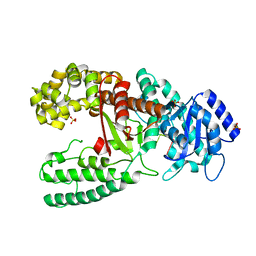 | | CRYSTAL STRUCTURE OF THE COMPLEX OF DNA POLYMERASE I KLENOW FRAGMENT WITH DNA | | Descriptor: | D(T)19 OLIGOMER, DNA POLYMERASE I, SULFATE ION, ... | | Authors: | Teplova, M, Wallace, S.T, Tereshko, V, Minasov, G, Simons, A.M, Cook, P.D, Manoharan, M, Egli, M. | | Deposit date: | 1999-10-26 | | Release date: | 1999-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural origins of the exonuclease resistance of a zwitterionic RNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7P0H
 
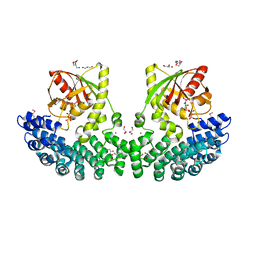 | | Crystal structure of Helicobacter pylori ComF fused to an artificial alphaREP crystallization helper(named B2) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, GLYCEROL, Helicobacter pylori ComF fused to an artificial alphaREP crystallization helper (named B2), ... | | Authors: | Celma, L, Walbott, H, Legrand, P, Quevillon-Cheruel, S. | | Deposit date: | 2021-06-29 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | ComFC mediates transport and handling of single-stranded DNA during natural transformation.
Nat Commun, 13, 2022
|
|
1CIV
 
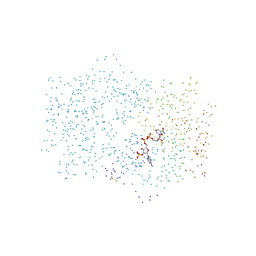 | | CHLOROPLAST NADP-DEPENDENT MALATE DEHYDROGENASE FROM FLAVERIA BIDENTIS | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-MALATE DEHYDROGENASE | | Authors: | Carr, P.D, Ashton, A.R, Verger, D, Ollis, D.L. | | Deposit date: | 1999-04-01 | | Release date: | 2000-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chloroplast NADP-malate dehydrogenase: structural basis of light-dependent regulation of activity by thiol oxidation and reduction.
Structure Fold.Des., 7, 1999
|
|
1CJA
 
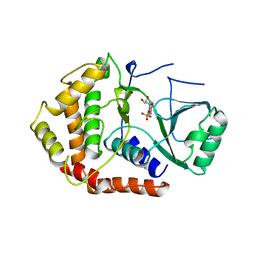 | | ACTIN-FRAGMIN KINASE, CATALYTIC DOMAIN FROM PHYSARUM POLYCEPHALUM | | Descriptor: | ADENOSINE MONOPHOSPHATE, PROTEIN (ACTIN-FRAGMIN KINASE) | | Authors: | Steinbacher, S, Hof, P, Eichinger, L, Schleicher, M, Gettemans, J, Vandekerckhove, J, Huber, R, Benz, J. | | Deposit date: | 1999-04-08 | | Release date: | 1999-06-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of the Physarum polycephalum actin-fragmin kinase: an atypical protein kinase with a specialized substrate-binding domain.
EMBO J., 18, 1999
|
|
1CR8
 
 | |
6OEI
 
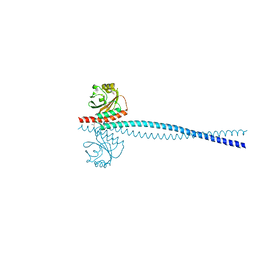 | | Yeast Spc42 N-terminal coiled-coil fused to PDB: 3K2N | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Spindle pole body component SPC42,Sigma-54-dependent transcriptional regulator | | Authors: | Drennan, A.C, Krishna, S, Seeger, M.A, Andreas, M.P, Gardner, J.M, Sether, E.K.R, Jaspersen, S.L, Rayment, I. | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure and function of Spc42 coiled-coils in yeast centrosome assembly and duplication.
Mol.Biol.Cell, 30, 2019
|
|
6OKB
 
 | | Prohead 2 of the phage T5 | | Descriptor: | Major capsid protein | | Authors: | Huet, A, Duda, R.L, Boulanger, P, Conway, J.F. | | Deposit date: | 2019-04-12 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Capsid expansion of bacteriophage T5 revealed by high resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1CGH
 
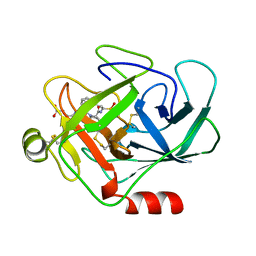 | | Human cathepsin G | | Descriptor: | CATHEPSIN G, N-(3-carboxypropanoyl)-L-valyl-N-{(1R)-1-[(S)-hydroxy(oxido)phosphanyl]-2-phenylethyl}-L-prolinamide | | Authors: | Hof, P, Bode, W. | | Deposit date: | 1996-06-26 | | Release date: | 1997-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A crystal structure of human cathepsin G in complex with Suc-Val-Pro-PheP-(OPh)2: a Janus-faced proteinase with two opposite specificities.
EMBO J., 15, 1996
|
|
8DEZ
 
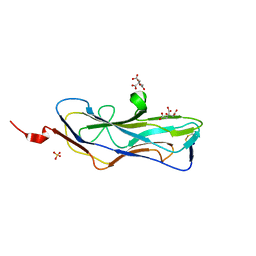 | | Abp2D Receptor Binding Domain ACICU | | Descriptor: | Abp2D Receptor Binding Domain, CITRATE ANION, SULFATE ION | | Authors: | Tamadonfar, K.O, Pinkner, J.P, Dodson, K.W, Kalas, V, Hultgren, S.J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structure-function correlates of fibrinogen binding by Acinetobacter adhesins critical in catheter-associated urinary tract infections.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1CLV
 
 | | YELLOW MEAL WORM ALPHA-AMYLASE IN COMPLEX WITH THE AMARANTH ALPHA-AMYLASE INHIBITOR | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN (ALPHA-AMYLASE INHIBITOR), ... | | Authors: | Pereira, P.J.B, Lozanov, V, Patthy, A, Huber, R, Bode, W, Pongor, S, Strobl, S. | | Deposit date: | 1999-05-04 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific inhibition of insect alpha-amylases: yellow meal worm alpha-amylase in complex with the amaranth alpha-amylase inhibitor at 2.0 A resolution.
Structure Fold.Des., 7, 1999
|
|
6O9Q
 
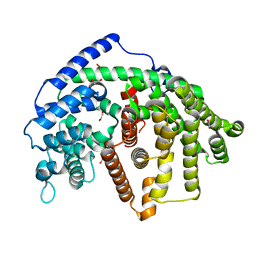 | | Wild-type SaSQS1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Sesquisabinene B synthase 1 | | Authors: | Blank, P.N, Christianson, D.W. | | Deposit date: | 2019-03-14 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Sesquisabinene Synthase 1, a Terpenoid Cyclase That Generates a Strained [3.1.0] Bridged-Bicyclic Product.
Acs Chem.Biol., 14, 2019
|
|
6O9P
 
 | | Wild-type SaSQS1 Complexed with Ibandronate | | Descriptor: | 1,2-ETHANEDIOL, IBANDRONATE, MAGNESIUM ION, ... | | Authors: | Blank, P.N, Christianson, D.W. | | Deposit date: | 2019-03-14 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Sesquisabinene Synthase 1, a Terpenoid Cyclase That Generates a Strained [3.1.0] Bridged-Bicyclic Product.
Acs Chem.Biol., 14, 2019
|
|
8CNX
 
 | | Structure of Enterovirus D68 3C protease | | Descriptor: | Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of EV D68 3C protease
To Be Published
|
|
5LYD
 
 | | Crystal structure of 1 in complex with tafCPB | | Descriptor: | (2~{S})-6-azanyl-2-(sulfamoylamino)hexanoic acid, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-09-27 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Sulfamide as Zinc Binding Motif in Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
J. Med. Chem., 59, 2016
|
|
