1ZH0
 
 | | Crystal Structure of L-3-(2-napthyl)alanine-tRNA synthetase in complex with L-3-(2-napthyl)alanine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-(2-NAPHTHYL)-ALANINE, Tyrosyl-tRNA synthetase | | Authors: | Turner, J.M, Graziano, J, Spraggon, G, Schultz, P.G. | | Deposit date: | 2005-04-22 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity of an aminoacyl-tRNA synthetase active site
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5T3N
 
 | | Sp-2Cl-cAMPS bound to PKAR CBD2 | | Descriptor: | (2S,4aR,6R,7R,7aS)-6-(6-amino-2-chloro-9H-purin-9-yl)-7-hydroxy-2-sulfanyltetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-2-one, IODIDE ION, cAMP-dependent protein kinase regulatory subunit | | Authors: | Littler, D.R, Gilson, P. | | Deposit date: | 2016-08-26 | | Release date: | 2016-10-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Disrupting the Allosteric Interaction between the Plasmodium falciparum cAMP-dependent Kinase and Its Regulatory Subunit.
J. Biol. Chem., 291, 2016
|
|
5T1X
 
 | | Crystal Structure of Native Tarin Lectin | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lectin, ... | | Authors: | Pereira, P.R, Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2016-08-22 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structures of Colocasia esculenta tarin lectin.
Glycobiology, 27, 2017
|
|
1PE8
 
 | | Thermolysin with monocyclic inhibitor | | Descriptor: | 2-ETHOXYETHYLPHOSPHINATE, 3-METHYLBUTAN-1-AMINE, CALCIUM ION, ... | | Authors: | Juers, D, Pyun, H.-J, Bartlett, P.A, Matthews, B.W. | | Deposit date: | 2003-05-21 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational Constraint and Structural Complementarity in Thermolysin Inhibitors: Structures of Enzyme Complexes and Conclusions
To be Published
|
|
4LGX
 
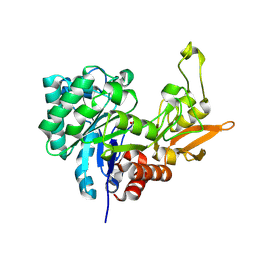 | | Structure of Chitinase D from Serratia proteamaculans revealed an unusually constrained substrate binding site | | Descriptor: | ACETATE ION, GLYCEROL, Glycoside hydrolase family 18 | | Authors: | Madhuprakash, J, Singh, A, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Podile, A.R, Singh, T.P. | | Deposit date: | 2013-06-30 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Inverse relationship between chitobiase and transglycosylation activities of chitinase-D from Serratia proteamaculans revealed by mutational and biophysical analyses.
Sci Rep, 5, 2015
|
|
4HFD
 
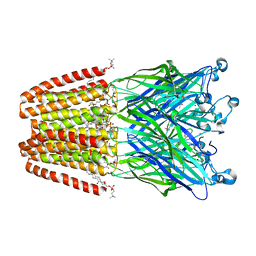 | | The GLIC pentameric Ligand-Gated Ion Channel F14'A ethanol-sensitive mutant complexed to bromoform | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Howard, R.J, Malherbe, L, Lee, U.S, Corringer, P.J, Harris, R.A, Delarue, M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for potentiation by alcohols and anaesthetics in a ligand-gated ion channel.
Nat Commun, 4, 2013
|
|
2NXY
 
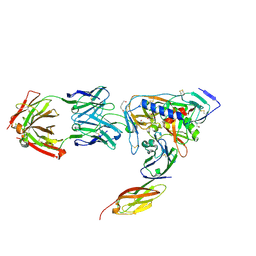 | | HIV-1 gp120 Envelope Glycoprotein(S334A) Complexed with CD4 and Antibody 17b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, ... | | Authors: | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
3GRU
 
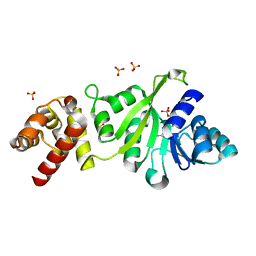 | |
2O0B
 
 | |
1FHT
 
 | | RNA-BINDING DOMAIN OF THE U1A SPLICEOSOMAL PROTEIN U1A117, NMR, 43 STRUCTURES | | Descriptor: | U1 SMALL NUCLEAR RIBONUCLEOPROTEIN A | | Authors: | Allain, F.H.-T, Gubser, C.C, Howe, P.W.A, Nagai, K, Neuhaus, D, Varani, G. | | Deposit date: | 1996-02-21 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding.
J.Mol.Biol., 257, 1996
|
|
2H2K
 
 | | Crystal Structure Analysis of Human S100A13 | | Descriptor: | CALCIUM ION, Protein S100-A13 | | Authors: | Li, M, Zhang, P.F, Zhang, J.P, Chang, W.R. | | Deposit date: | 2006-05-19 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure study on human S100A13 at 2.0 A resolution
Biochem.Biophys.Res.Commun., 356, 2007
|
|
1OVG
 
 | | M64V PNP +MePdr | | Descriptor: | 9-(2-DEOXY-BETA-D-RIBOFURANOSYL)-6-METHYLPURINE, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Ealick, S.E, Bennett, E.M, Anand, R, Secrist, J.A, Parker, P.W, Hassan, A.E, Allan, P.W, McPherson, D.T, Sorscher, E.J. | | Deposit date: | 2003-03-26 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Designer gene therapy using an Escherichia coli purine nucleoside phosphorylase/prodrug system.
Chem.Biol., 10, 2003
|
|
7DUQ
 
 | | Cryo-EM structure of the compound 2 and GLP-1-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1, Glucagon-like peptide 1 receptor, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
1FNB
 
 | | REFINED CRYSTAL STRUCTURE OF SPINACH FERREDOXIN REDUCTASE AT 1.7 ANGSTROMS RESOLUTION: OXIDIZED, REDUCED, AND 2'-PHOSPHO-5'-AMP BOUND STATES | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Bruns, C.M, Karplus, P.A. | | Deposit date: | 1995-01-05 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined crystal structure of spinach ferredoxin reductase at 1.7 A resolution: oxidized, reduced and 2'-phospho-5'-AMP bound states.
J.Mol.Biol., 247, 1995
|
|
5BP3
 
 | | Dehydratase domain (DH) of a mycocerosic acid synthase-like (MAS-like) PKS, crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, Mycocerosic acid synthase-like polyketide synthase | | Authors: | Herbst, D.A, Jakob, P.R, Zaehringer, F, Maier, T. | | Deposit date: | 2015-05-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Mycocerosic acid synthase exemplifies the architecture of reducing polyketide synthases.
Nature, 531, 2016
|
|
4LCG
 
 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-050 complex | | Descriptor: | (betaS)-Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N,beta-dihydroxy-L-tyrosinamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2013-06-21 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.568 Å) | | Cite: | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
1P5P
 
 | |
3GYR
 
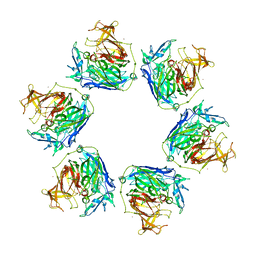 | | Structure of Phenoxazinone synthase from Streptomyces antibioticus reveals a new type 2 copper center. | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, GLYCEROL, ... | | Authors: | Smith, A.W, Camara-Artigas, A, Wang, M, Francisco, W.A, Allen, J.P. | | Deposit date: | 2009-04-05 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Phenoxazinone Synthase from Streptomyces Antibioticus Reveals a New Type 2 Copper Center.
Biochemistry, 45, 2006
|
|
4KWM
 
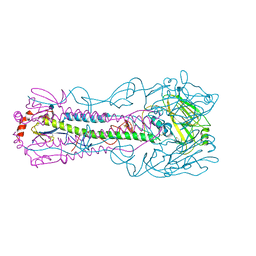 | | Structure of a/anhui/5/2005 h5 ha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Shore, D.A, Carney, P.J, Chang, J.C, Stevens, J. | | Deposit date: | 2013-05-24 | | Release date: | 2014-06-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and antigenic variation among diverse clade 2 H5N1 viruses.
Plos One, 8, 2013
|
|
4KXM
 
 | |
4KY8
 
 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, methotrexate, FdUMP and 4-((2-amino-6-methyl-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)thio)-2-chlorophenyl)-L-glutamic acid | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional thymidylate synthase-dihydrofolate reductase, METHOTREXATE, ... | | Authors: | Kumar, V.P, Anderson, K.S. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.084 Å) | | Cite: | Substituted pyrrolo[2,3-d]pyrimidines as Cryptosporidium hominis thymidylate synthase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1P4G
 
 | | Crystal structure of glycogen phosphorylase b in complex with C-(1-azido-alpha-D-glucopyranosyl)formamide | | Descriptor: | C-(1-AZIDO-ALPHA-D-GLUCOPYRANOSYL) FORMAMIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Oikonomakos, N.G, Zographos, S.E, Kosmopoulou, M.N, Bischler, N, Leonidas, D.D, Nagy, V, Somsak, L, Gergely, P, Praly, J.-P. | | Deposit date: | 2003-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crsytallographic Studies on alpha- and beta-D-glucopyranosyl Formamide Analogues, Inhibitors of Glycogen Phosphorylase
Biocatal.Biotransfor., 21, 2003
|
|
1FZX
 
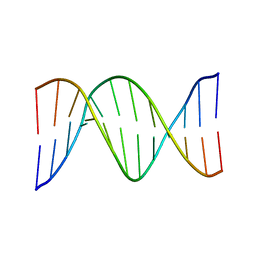 | | NMR SOLUTION STRUCTURE OF THE DNA DODECAMER GGCAAAAAACGG | | Descriptor: | 5'-D(*CP*CP*GP*TP*TP*TP*TP*TP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*AP*AP*AP*AP*AP*CP*GP*G)-3' | | Authors: | MacDonald, D, Herbert, K, Zhang, X, Pologruto, T, Lu, P. | | Deposit date: | 2000-10-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an A-tract DNA bend.
J.Mol.Biol., 306, 2001
|
|
3IWP
 
 | |
4G4J
 
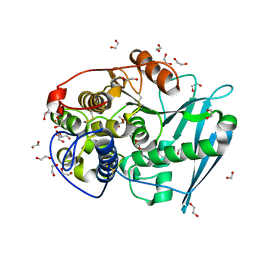 | | Crystal structure of glucuronoyl esterase S213A mutant from Sporotrichum thermophile in complex with methyl 4-O-methyl-beta-D-glucopyranuronate determined at 2.35 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-glucuronoyl methylesterase, GLYCEROL, ... | | Authors: | Charavgi, M.D, Dimarogona, M, Topakas, E, Christakopoulos, P, Chrysina, E.D. | | Deposit date: | 2012-07-16 | | Release date: | 2013-01-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of a novel glucuronoyl esterase from Myceliophthora thermophila gives new insights into its role as a potential biocatalyst.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
