2W92
 
 | | Structure of a Streptococcus pneumoniae family 85 glycoside hydrolase, Endo-D, in complex with NAG-thiazoline. | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ENDO-BETA-N-ACETYLGLUCOSAMINIDASE D, PENTAETHYLENE GLYCOL | | Authors: | Abbott, D.W, Macauley, M.S, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2009-01-21 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Streptococcus Pneumoniae Endohexosaminidase D, Structural and Mechanistic Insight Into Substrate-Assisted Catalysis in Family 85 Glycoside Hydrolases.
J.Biol.Chem., 284, 2009
|
|
2XD3
 
 | | The crystal structure of MalX from Streptococcus pneumoniae in complex with maltopentaose. | | Descriptor: | MALTOSE/MALTODEXTRIN-BINDING PROTEIN, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Abbott, D.W, Higgins, M.A, Hyrnuik, S, Pluvinage, B, Lammerts van Bueren, A, Boraston, A.B. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Molecular Basis of Glycogen Breakdown and Transport in Streptococcus Pneumoniae.
Mol.Microbiol., 77, 2010
|
|
2XJQ
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae | | Descriptor: | CHLORIDE ION, FLOCCULATION PROTEIN FLO5, GLYCEROL, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1TD4
 
 | | Crystal structure of VSHP_BPP21 in space group H3 with high resolution. | | Descriptor: | Head decoration protein | | Authors: | Chang, C, Forrer, P, Ott, D, Wlodawer, A, Plueckthun, A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kinetic Stability and Crystal Structure of the Viral Capsid Protein SHP.
J.Mol.Biol., 344, 2004
|
|
1TD3
 
 | | Crystal structure of VSHP_BPP21 in space group C2 | | Descriptor: | Head decoration protein | | Authors: | Chang, C, Forrer, P, Ott, D, Wlodawer, A, Plueckthun, A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Kinetic Stability and Crystal Structure of the Viral Capsid Protein SHP.
J.Mol.Biol., 344, 2004
|
|
1TD0
 
 | | Viral capsid protein SHP at pH 5.5 | | Descriptor: | Head decoration protein | | Authors: | Chang, C, Forrer, P, Ott, D, Wlodawer, A, Plueckthun, A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Kinetic Stability and Crystal Structure of the Viral Capsid Protein SHP
J.Mol.Biol., 344, 2004
|
|
2XJV
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae with mutation D201T in complex with calcium and glucose | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XJS
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae in complex with calcium and a1,2-mannobiose | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XJU
 
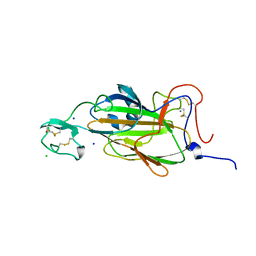 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae with mutation S227A in complex with calcium and a1,2-mannobiose | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XJR
 
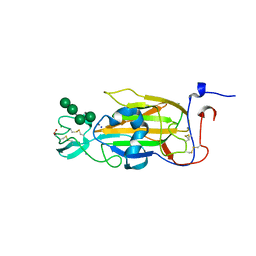 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae in complex with calcium and Man5(D2-D3) | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XJT
 
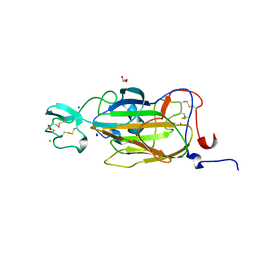 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae in complex with calcium and Man5(D1) | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XJP
 
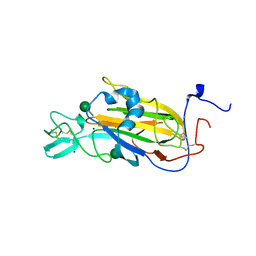 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae in complex with calcium and mannose | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6P5W
 
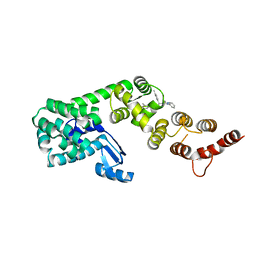 | | Structure of DCN1 bound to 3-methyl-N-((4S,5S)-3-methyl-6-oxo-1-phenyl-4-(p-tolyl)-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)benzamide | | Descriptor: | 3-methyl-N-[(4S,5S)-3-methyl-4-(4-methylphenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]benzamide, Lysozyme,DCN1-like protein 1 chimera | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
6V4T
 
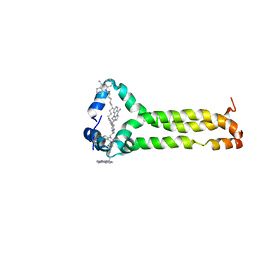 | | MPER-TMD of HIV-1 Env bound with the entry inhibitor S2C3 | | Descriptor: | 4,4'-(decane-1,10-diyl)bis(9-amino-2,3-dihydro-1H-cyclopenta[b]quinolin-4-ium), Envelope glycoprotein gp160 | | Authors: | Xiao, T, Frey, G, Fu, Q, Lavine, C.L, Scott, D.A, Seaman, M.S, Chou, J.J, Chen, B. | | Deposit date: | 2019-11-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HIV-1 fusion inhibitors targeting the membrane-proximal external region of Env spikes.
Nat.Chem.Biol., 16, 2020
|
|
6VRE
 
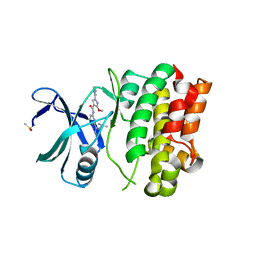 | | Structure of ASK1 bound to BIO-1772961 | | Descriptor: | 3-methoxy-N-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}-1-(pyrazin-2-yl)-1H-pyrazole-4-carboxamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Chodaparambil, J.V, Marcotte, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of CNS-Penetrant Apoptosis Signal-Regulating Kinase 1 (ASK1) Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6P5V
 
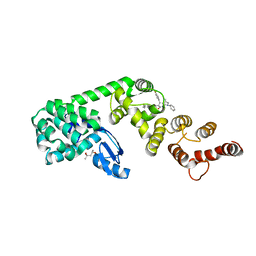 | | Structure of DCN1 bound to N-((4S,5S)-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-methylbenzamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lysozyme,DCN1-like protein 1 fusion, N-[(4S,5S)-1-[(1S)-cyclohex-3-en-1-yl]-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-methylbenzamide | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
6VRF
 
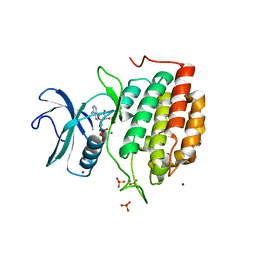 | | ADP bound TTBK2 kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chodaparambil, J.V, Marcotte, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanisms of Regulation and Diverse Activities of Tau-Tubulin Kinase (TTBK) Isoforms.
Cell Mol Neurobiol, 41, 2021
|
|
4UQO
 
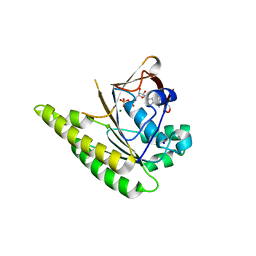 | | RADA C-TERMINAL ATPASE DOMAIN FROM PYROCOCCUS FURIOSUS BOUND TO ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA REPAIR AND RECOMBINATION PROTEIN RADA, MAGNESIUM ION, ... | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2014-06-24 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
2VMH
 
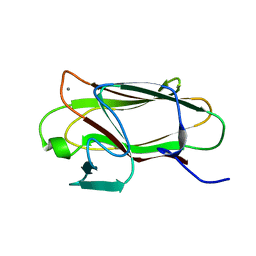 | | The structure of CBM51 from Clostridium perfringens GH95 | | Descriptor: | CALCIUM ION, FIBRONECTIN TYPE III DOMAIN PROTEIN | | Authors: | Gregg, K, Finn, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2008-01-25 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Divergent Modes of Glycan Recognition by a New Family of Carbohydrate-Binding Modules
J.Biol.Chem., 283, 2008
|
|
2VMI
 
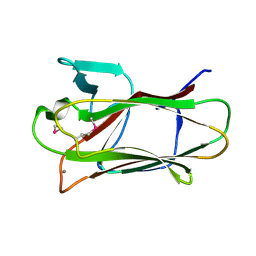 | | The structure of seleno-methionine labelled CBM51 from Clostridium perfringens GH95 | | Descriptor: | CALCIUM ION, FIBRONECTIN TYPE III DOMAIN PROTEIN | | Authors: | Gregg, K, Finn, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2008-01-25 | | Release date: | 2008-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Divergent Modes of Glycan Recognition by a New Family of Carbohydrate-Binding Modules
J.Biol.Chem., 283, 2008
|
|
6W07
 
 | | Bruton's tyrosine kinase in complex with compound 1 | | Descriptor: | DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK, ~{N}-[[2-methyl-4-[2-[(1-methylpyrazol-4-yl)amino]pyrimidin-4-yl]phenyl]methyl]-3-propan-2-yloxy-azetidine-1-carboxamide | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Discovery of BIIB068: A Selective, Potent, Reversible Bruton's Tyrosine Kinase Inhibitor as an Orally Efficacious Agent for Autoimmune Diseases.
J.Med.Chem., 63, 2020
|
|
5JZL
 
 | | The Structure of Monomeric Ultra Stable Green Fluorescent Protein | | Descriptor: | CHLORIDE ION, Green fluorescent protein, SODIUM ION | | Authors: | Gunn, N.J, Yong, K.J, Scott, D.J, Griffin, M.D.W. | | Deposit date: | 2016-05-17 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel Ultra-Stable, Monomeric Green Fluorescent Protein For Direct Volumetric Imaging of Whole Organs Using CLARITY.
Sci Rep, 8, 2018
|
|
4GBA
 
 | | DCNL complex with N-terminally acetylated NEDD8 E2 peptide | | Descriptor: | DCN1-like protein 3, NEDD8-conjugating enzyme UBE2F | | Authors: | Monda, J.K, Scott, D.C, Miller, D.J, Harper, J.W, Bennett, E.J, Schulman, B.A. | | Deposit date: | 2012-07-26 | | Release date: | 2012-11-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Conservation of Distinctive N-terminal Acetylation-Dependent Interactions across a Family of Mammalian NEDD8 Ligation Enzymes.
Structure, 21, 2013
|
|
6PB0
 
 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
2VMG
 
 | | The structure of CBM51 from Clostridium perfringens GH95 in complex with methyl-galactose | | Descriptor: | CALCIUM ION, FIBRONECTIN TYPE III DOMAIN PROTEIN, methyl beta-D-galactopyranoside | | Authors: | Gregg, K, Finn, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2008-01-25 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Divergent Modes of Glycan Recognition by a New Family of Carbohydrate-Binding Modules
J.Biol.Chem., 283, 2008
|
|
