1TYK
 
 | | SOLUTION STRUCTURE OF A TOXIN FROM THE TARANTULA, GRAMMOSTOLA SPATULATA, WHICH INHIBITS MECHANOSENSITIVE ION CHANNELS | | 分子名称: | Toxin GsMTx-4 | | 著者 | Oswald, R.E, Suchyna, T.M, Mcfeeters, R, Gottlieb, P, Sachs, F. | | 登録日 | 2004-07-08 | | 公開日 | 2004-07-13 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Peptide Toxins that Block Mechanosensitive Ion Channels
J.Biol.Chem., 277, 2002
|
|
2NBT
 
 | | NEURONAL BUNGAROTOXIN, NMR, 10 STRUCTURES | | 分子名称: | NEURONAL BUNGAROTOXIN | | 著者 | Oswald, R.E, Sutcliffe, M.J, Bamberger, M, Loring, R.H, Braswell, E, Dobson, C.M. | | 登録日 | 1997-10-29 | | 公開日 | 1998-03-11 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of neuronal bungarotoxin determined by two-dimensional NMR spectroscopy: calculation of tertiary structure using systematic homologous model building, dynamical simulated annealing, and restrained molecular dynamics.
Biochemistry, 31, 1992
|
|
1LUP
 
 | | Solution structure of a toxin (GsMTx2) from the tarantula, Grammostola spatulata, which inhibits mechanosensitive ion channels | | 分子名称: | GsMTx2 | | 著者 | Oswald, R.E, Suchyna, T.M, McFeeters, R, Gottlieb, P, Sachs, F. | | 登録日 | 2002-05-23 | | 公開日 | 2002-08-07 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of peptide toxins that block mechanosensitive ion channels
J.Biol.Chem., 277, 2002
|
|
1EES
 
 | | SOLUTION STRUCTURE OF CDC42HS COMPLEXED WITH A PEPTIDE DERIVED FROM P-21 ACTIVATED KINASE, NMR, 20 STRUCTURES | | 分子名称: | GTP-BINDING PROTEIN, P21-ACTIVATED KINASE | | 著者 | Gizachew, D, Guo, W, Chohan, K.C, Sutcliffe, M.J, Oswald, R.E. | | 登録日 | 2000-02-02 | | 公開日 | 2000-03-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the complex of Cdc42Hs with a peptide derived from P-21 activated kinase.
Biochemistry, 39, 2000
|
|
2MOG
 
 | | Solution structure of the terminal Ig-like domain from Leptospira interrogans LigB | | 分子名称: | Bacterial Ig-like domain, group 2 | | 著者 | Ptak, C.P, Hsieh, C, Lin, Y, Maltsev, A.S, Raman, R, Sharma, Y, Oswald, R.E, Chang, Y. | | 登録日 | 2014-04-25 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Solution Structure of the Terminal Immunoglobulin-like Domain from the Leptospira Host-Interacting Outer Membrane Protein, LigB.
Biochemistry, 53, 2014
|
|
2ASE
 
 | |
3LSF
 
 | | Piracetam bound to the ligand binding domain of GluA2 | | 分子名称: | 2-(2-oxopyrrolidin-1-yl)acetamide, GLUTAMIC ACID, Glutamate receptor 2, ... | | 著者 | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | 登録日 | 2010-02-12 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.851 Å) | | 主引用文献 | Piracetam Defines a New Binding Site for Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazole-propionic Acid (AMPA) Receptors.
J.Med.Chem., 53, 2010
|
|
3LSL
 
 | |
3M3F
 
 | | PEPA bound to the ligand binding domain of GluA3 (flop form) | | 分子名称: | 2-[2,6-difluoro-4-({2-[(phenylsulfonyl)amino]ethyl}sulfanyl)phenoxy]acetamide, GLUTAMIC ACID, Glutamate receptor 3, ... | | 著者 | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | 登録日 | 2010-03-09 | | 公開日 | 2010-03-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular mechanism of flop selectivity and subsite recognition for an AMPA receptor allosteric modulator: structures of GluA2 and GluA3 in complexes with PEPA.
Biochemistry, 49, 2010
|
|
1AJE
 
 | | CDC42 FROM HUMAN, NMR, 20 STRUCTURES | | 分子名称: | CDC42HS | | 著者 | Feltham, J.L, Dotsch, V, Raza, S, Manor, D, Cerione, R.A, Sutcliffe, M.J, Wagner, G, Oswald, R.E. | | 登録日 | 1997-05-02 | | 公開日 | 1997-11-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Definition of the switch surface in the solution structure of Cdc42Hs.
Biochemistry, 36, 1997
|
|
2KB0
 
 | |
4Q30
 
 | |
4GXS
 
 | | Ligand binding domain of GluA2 (AMPA/glutamate receptor) bound to (-)-kaitocephalin | | 分子名称: | (5R)-2-[(1S,2R)-2-amino-2-carboxy-1-hydroxyethyl]-5-{(2S)-2-carboxy-2-[(3,5-dichloro-4-hydroxybenzoyl)amino]ethyl}-L-proline, Glutamate receptor 2, ZINC ION | | 著者 | Ahmed, A.H, Oswald, R.E. | | 登録日 | 2012-09-04 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9634 Å) | | 主引用文献 | The structure of (-)-kaitocephalin bound to the ligand binding domain of the (S)-alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)/glutamate receptor, GluA2.
J.Biol.Chem., 287, 2012
|
|
7TOD
 
 | | Solution structure of the phosphatidylinositol 3-phosphate binding domain from the Legionella effector SetA | | 分子名称: | Subversion of eukaryotic traffic protein A | | 著者 | Beck, W.H.J, Enoki, T.A, Feigenson, G.W, Nicholson, L, Oswald, R, Mao, Y. | | 登録日 | 2022-01-24 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the phosphatidylinositol 3-phosphate binding domain from the Legionella effector SetA
To Be Published
|
|
3M3K
 
 | | Ligand binding domain (S1S2) of GluA3 (flop) | | 分子名称: | GLUTAMIC ACID, Glutamate receptor 3, ZINC ION | | 著者 | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | 登録日 | 2010-03-09 | | 公開日 | 2010-03-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.793 Å) | | 主引用文献 | Molecular mechanism of flop selectivity and subsite recognition for an AMPA receptor allosteric modulator: structures of GluA2 and GluA3 in complexes with PEPA.
Biochemistry, 49, 2010
|
|
3M3L
 
 | | PEPA bound to the ligand binding domain of GluA2 (flop form) | | 分子名称: | 2-[2,6-difluoro-4-({2-[(phenylsulfonyl)amino]ethyl}sulfanyl)phenoxy]acetamide, GLUTAMIC ACID, Glutamate receptor 2, ... | | 著者 | Ptak, C.P, Ahmed, A.H, Oswald, R.E. | | 登録日 | 2010-03-09 | | 公開日 | 2010-03-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Molecular mechanism of flop selectivity and subsite recognition for an AMPA receptor allosteric modulator: structures of GluA2 and GluA3 in complexes with PEPA.
Biochemistry, 49, 2010
|
|
4F22
 
 | |
3T9U
 
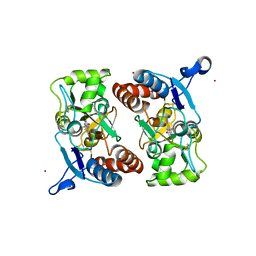 | |
3IL1
 
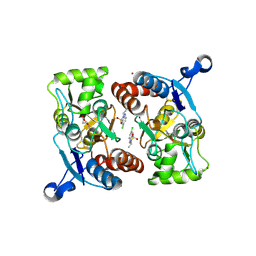 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, IDRA-21 | | 分子名称: | (3S)-7-chloro-3-methyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | 著者 | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | 登録日 | 2009-08-06 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3ILT
 
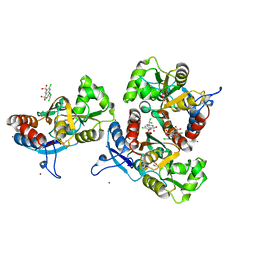 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, trichlormethiazide | | 分子名称: | 6-CHLORO-3-(DICHLOROMETHYL)-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE-7-SULFONAMIDE 1,1-DIOXIDE, GLUTAMIC ACID, Glutamate receptor 2, ... | | 著者 | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | 登録日 | 2009-08-07 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.107 Å) | | 主引用文献 | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3IK6
 
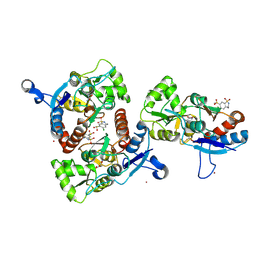 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, chlorothiazide | | 分子名称: | 6-chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | 著者 | Ptak, C.P, Ahmed, A.H, Oswald, R.E. | | 登録日 | 2009-08-05 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3IJO
 
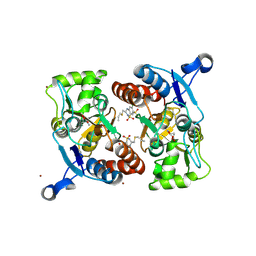 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, althiazide | | 分子名称: | (3S)-6-chloro-3-[(prop-2-en-1-ylsulfanyl)methyl]-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | 著者 | Ptak, C.P, Ahmed, A.H, Oswald, R.E. | | 登録日 | 2009-08-04 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.003 Å) | | 主引用文献 | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3IJX
 
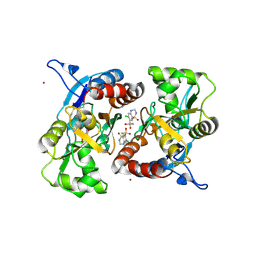 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, hydrochlorothiazide | | 分子名称: | 6-chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | 著者 | Ptak, C.P, Ahmed, A.H, Oswald, R.E. | | 登録日 | 2009-08-05 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.881 Å) | | 主引用文献 | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3ILU
 
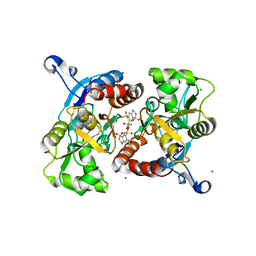 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, hydroflumethiazide | | 分子名称: | 6-(trifluoromethyl)-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | 著者 | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | 登録日 | 2009-08-07 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.003 Å) | | 主引用文献 | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3LSW
 
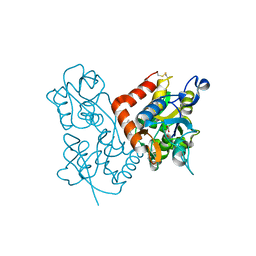 | | Aniracetam bound to the ligand binding domain of GluA3 | | 分子名称: | 1-(4-METHOXYBENZOYL)-2-PYRROLIDINONE, GLUTAMIC ACID, GluA2 S1S2 domain, ... | | 著者 | Ahmed, A.H, Oswald, R.E. | | 登録日 | 2010-02-13 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.752 Å) | | 主引用文献 | Piracetam Defines a New Binding Site for Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazole-propionic Acid (AMPA) Receptors.
J.Med.Chem., 53, 2010
|
|
