2P0N
 
 | | NMB1532 protein from Neisseria meningitidis, unknown function | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Osipiuk, J, Li, H, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | X-ray crystal structure of NMB1532 protein of unknown function from Neisseria meningitidis.
To be Published
|
|
1Q77
 
 | | X-ray crystal structure of putative Universal Stress Protein from Aquifex aeolicus | | Descriptor: | Hypothetical protein AQ_178, SULFATE ION | | Authors: | Osipiuk, J, Zhou, M, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-16 | | Release date: | 2003-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural homolog of Universal Stress Protein from Aquifex aeolicus
To be Published
|
|
6XG3
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, at room temperature | | Descriptor: | CHLORIDE ION, Non-structural protein 3, PHOSPHATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
5E7Q
 
 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis | | Descriptor: | GLYCEROL, SULFATE ION, acyl-CoA synthetase | | Authors: | Osipiuk, J, Cuff, M.E, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J, Ma, M, Chang, C.Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-12 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
5DUL
 
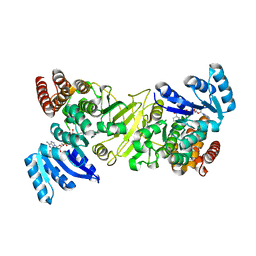 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Osipiuk, J, Mulligan, R, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH .
to be published
|
|
8G62
 
 | | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004 | | Descriptor: | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Luci, D, Kales, S, Simeonov, A, Rai, G, Drayman, N, Tay, S, Oakes, S, Rosner, M, Chen, B, Dulin, N, Solway, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004
To Be Published
|
|
1HJO
 
 | | ATPase domain of human heat shock 70kDa protein 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Walsh, M.A, Freeman, B.C, Morimoto, R.I, Joachimiak, A. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a new crystal form of human Hsp70 ATPase domain.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5DGG
 
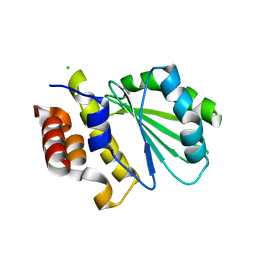 | | Central domain of uncharacterized Lpg1148 protein from Legionella pneumophila | | Descriptor: | CHLORIDE ION, Uncharacterized protein | | Authors: | Osipiuk, J, Evdokimova, E, Yim, V, Joachimiak, A, Ensminger, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol. Syst. Biol., 12, 2016
|
|
8T28
 
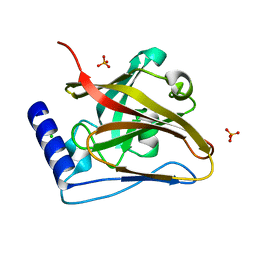 | | The crystal structure of SrtC2 sortase from Actinomyces oris | | Descriptor: | CHLORIDE ION, Class C sortase, PHOSPHATE ION | | Authors: | Osipiuk, J, Chang, C, Ton-That, H.L, Ton-That, H, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-06-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for dual functions in pilus assembly modulated by the lid of a pilus-specific sortase.
J.Biol.Chem., 300, 2024
|
|
8UW6
 
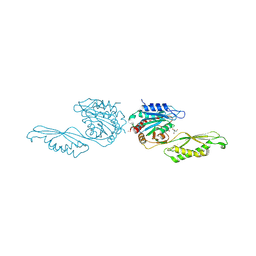 | | Acetylornithine deacetylase from Escherichia coli, di-zinc form. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acetylornithine deacetylase, ... | | Authors: | Osipiuk, J, Endres, M, Kelley, E, Becker, D.P, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-11-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N alpha-acetyl-L-ornithine deacetylase from Escherichia coli and a ninhydrin-based assay to enable inhibitor identification.
Front Chem, 12, 2024
|
|
6DFU
 
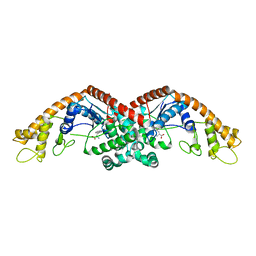 | | Tryptophan--tRNA ligase from Haemophilus influenzae. | | Descriptor: | TRYPTOPHAN, Tryptophan--tRNA ligase | | Authors: | Osipiuk, J, Maltseva, N, Mulligan, R, Grimshaw, S, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tryptophan--tRNA ligase from Haemophilus influenzae.
to be published
|
|
3NJH
 
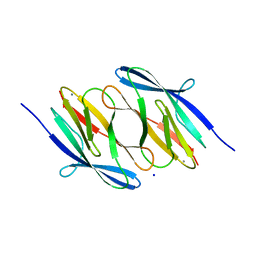 | | D37A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis. | | Descriptor: | CALCIUM ION, GLYCEROL, Peptidase, ... | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
4GL0
 
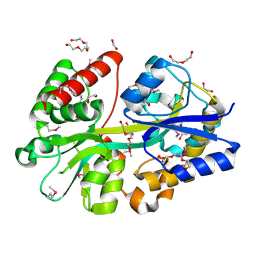 | | Putative spermidine/putrescine ABC transporter from Listeria monocytogenes | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Lmo0810 protein, ... | | Authors: | Osipiuk, J, Zhou, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-13 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Putative spermidine/putrescine ABC transporter from Listeria monocytogenes
To be Published
|
|
3N55
 
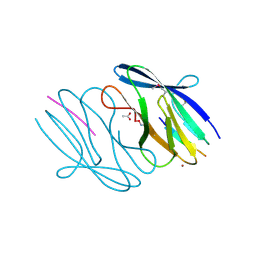 | | SO1698 protein, an aspartic peptidase from Shewanella oneidensis. | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, Peptidase, ... | | Authors: | Osipiuk, J, Mulligan, R, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-24 | | Release date: | 2010-06-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
4GCN
 
 | | N-terminal domain of stress-induced protein-1 (STI-1) from C.elegans | | Descriptor: | Protein STI-1, TRIETHYLENE GLYCOL | | Authors: | Osipiuk, J, Bigelow, L, Gu, M, Van Oosten-Hawle, P, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-30 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N-terminal domain of stress-induced protein-1 (STI-1) from C.elegans
To be Published
|
|
4GB7
 
 | | Putative 6-aminohexanoate-dimer hydrolase from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, 6-aminohexanoate-dimer hydrolase, NITRATE ION | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-07-26 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Putative 6-aminohexanoate-dimer hydrolase from Bacillus anthracis.
To be Published
|
|
4FXS
 
 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae complexed with IMP and mycophenolic acid | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, MYCOPHENOLIC ACID, ... | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-07-03 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae complexed with IMP and mycophenolic acid.
To be Published
|
|
3N4J
 
 | | Putative RNA methyltransferase from Yersinia pestis | | Descriptor: | RNA methyltransferase, SULFATE ION | | Authors: | Osipiuk, J, Maltseva, N, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | X-ray crystal structure of putative RNA methyltransferase from Yersinia pestis.
To be Published
|
|
3N4K
 
 | | Putative RNA methyltransferase from Yersinia pestis in complex with S-ADENOSYL-L-HOMOCYSTEINE. | | Descriptor: | RNA methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Osipiuk, J, Maltseva, N, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray crystal structure of putative RNA methyltransferase from Yersinia pestis.
To be Published
|
|
1G2R
 
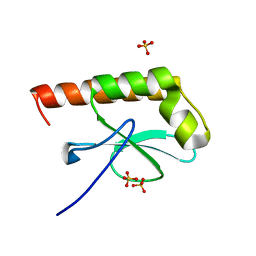 | | Structure of Cytosolic Protein of Unknown Function Coded by Gene from NUSA/INFB Region, a YlxR Homologue | | Descriptor: | HYPOTHETICAL CYTOSOLIC PROTEIN, SULFATE ION | | Authors: | Osipiuk, J, Gornicki, P, Maj, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-10-20 | | Release date: | 2001-08-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Streptococcus pneumonia YlxR at 1.35 A shows a putative new fold.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G60
 
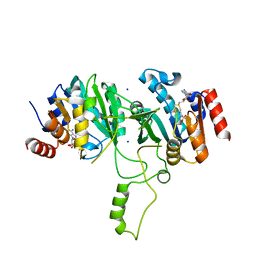 | | Crystal Structure of Methyltransferase MboIIa (Moraxella bovis) | | Descriptor: | Adenine-specific Methyltransferase MboIIA, S-ADENOSYLMETHIONINE, SODIUM ION | | Authors: | Osipiuk, J, Walsh, M.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-11-02 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of MboIIA methyltransferase.
Nucleic Acids Res., 31, 2003
|
|
6U0I
 
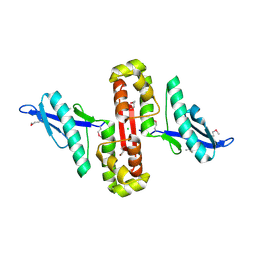 | |
3I1I
 
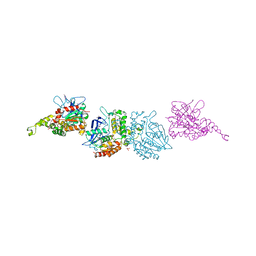 | | X-ray crystal structure of homoserine O-acetyltransferase from Bacillus anthracis | | Descriptor: | ACETATE ION, GLYCEROL, Homoserine O-acetyltransferase, ... | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | X-ray crystal structure of homoserine O-acetyltransferase from Bacillus anthracis.
To be published
|
|
3IC5
 
 | |
3IR9
 
 | |
