5KCN
 
 | |
5W0K
 
 | |
5VBG
 
 | |
5T1D
 
 | | Crystal structure of EBV gHgL/gp42/E1D1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, E1D1 IgG2a heavy chain, E1D1 IgG2a light chain, ... | | Authors: | Sathiyamoorthy, K, Jardetzky, T.S. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for Epstein-Barr virus host cell tropism mediated by gp42 and gHgL entry glycoproteins.
Nat Commun, 7, 2016
|
|
8DML
 
 | | Vibrio parahaemolyticus VtrA/VtrC complex bound to the bile salt chenodeoxycholate | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHENODEOXYCHOLIC ACID, ... | | Authors: | Tomchick, D.R, Orth, K, Zou, A.J. | | Deposit date: | 2022-07-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Molecular determinants for differential activation of the bile acid receptor from the pathogen Vibrio parahaemolyticus.
J.Biol.Chem., 299, 2023
|
|
6PE4
 
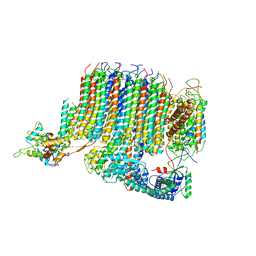 | | Yeast Vo motor in complex with 1 VopQ molecule | | Descriptor: | Cation transporter, Uncharacterized protein YPR170W-B, V-type proton ATPase subunit a, ... | | Authors: | Peng, W, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2019-06-20 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A distinct inhibitory mechanism of the V-ATPase by Vibrio VopQ revealed by cryo-EM.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6PE5
 
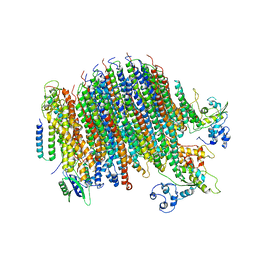 | | Yeast Vo motor in complex with 2 VopQ molecules | | Descriptor: | Cation transporter, Uncharacterized protein YPR170W-B, V-type proton ATPase subunit a, ... | | Authors: | Peng, W, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2019-06-20 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A distinct inhibitory mechanism of the V-ATPase by Vibrio VopQ revealed by cryo-EM.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8TWS
 
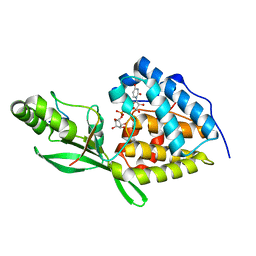 | | AvrB bound with UDP-rhamnose and RIN4 C-NOI motif | | Descriptor: | Avirulence protein B, RPM1-interacting protein 4, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{R},6~{S})-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Peng, W, Orth, K. | | Deposit date: | 2023-08-21 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Pseudomonas effector AvrB is a glycosyltransferase that rhamnosylates plant guardee protein RIN4.
Sci Adv, 10, 2024
|
|
8TXF
 
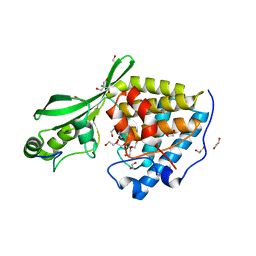 | | AvrB bound with RIN4 C-NOI motif | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Avirulence protein B, ... | | Authors: | Peng, W, Orth, K. | | Deposit date: | 2023-08-23 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Pseudomonas effector AvrB is a glycosyltransferase that rhamnosylates plant guardee protein RIN4.
Sci Adv, 10, 2024
|
|
8TWJ
 
 | | AvrB_R266A bound with UDP | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Avirulence protein B, ... | | Authors: | Peng, W, Orth, K. | | Deposit date: | 2023-08-21 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pseudomonas effector AvrB is a glycosyltransferase that rhamnosylates plant guardee protein RIN4.
Sci Adv, 10, 2024
|
|
8TWO
 
 | | AvrB bound with UDP and RIN4_T166-Rha | | Descriptor: | Avirulence protein B, RPM1-interacting protein 4, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Peng, W, Orth, K. | | Deposit date: | 2023-08-21 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Pseudomonas effector AvrB is a glycosyltransferase that rhamnosylates plant guardee protein RIN4.
Sci Adv, 10, 2024
|
|
6MRW
 
 | | 14-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
6MRT
 
 | | 12-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
6MRU
 
 | | 13-meric ClyA pore complex | | Descriptor: | Hemolysin E, chromosomal | | Authors: | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
5KEV
 
 | |
5KEW
 
 | | Vibrio parahaemolyticus VtrA/VtrC complex bound to the bile salt taurodeoxycholate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tomchick, D.R, Orth, K, Rivera-Cancel, G. | | Deposit date: | 2016-06-10 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Bile salt receptor complex activates a pathogenic type III secretion system.
Elife, 5, 2016
|
|
2OIV
 
 | | Structural Analysis of Xanthomonas XopD Provides Insights Into Substrate Specificity of Ubiquitin-like Protein Proteases | | Descriptor: | PHOSPHATE ION, Xanthomonas outer protein D | | Authors: | Chosed, R, Tomchick, D.R, Brautigam, C.A, Machius, M, Orth, K. | | Deposit date: | 2007-01-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases.
J.Biol.Chem., 282, 2007
|
|
2OIX
 
 | | Xanthomonas XopD C470A Mutant | | Descriptor: | Xanthomonas outer protein D | | Authors: | Chosed, R, Tomchick, D.R, Brautigam, C.A, Machius, M, Orth, K. | | Deposit date: | 2007-01-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases.
J.Biol.Chem., 282, 2007
|
|
3LET
 
 | | Crystal Structure of Fic domain containing AMPylator, VopS | | Descriptor: | Adenosine monophosphate-protein transferase vopS | | Authors: | Luong, P.H, Kinch, L.N, Brautigam, C.A, Grishin, N.V, Tomchick, D.R, Orth, K. | | Deposit date: | 2010-01-15 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural and Kinetic Analysis of VopS with Fic Domain Supports a Direct Transfer Mechanism for AMPylation
To be Published
|
|
5EGM
 
 | | Development of a novel tricyclic class of potent and selective FIXa inhibitors | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-chloranyl-~{N}-[(7~{S})-2-methyl-7-phenyl-10-(1~{H}-1,2,3,4-tetrazol-5-yl)-8,9-dihydro-6~{H}-pyrido[1,2-a]indol-7-yl]-4-(1,2,4-triazol-4-yl)benzamide, Coagulation factor IX, ... | | Authors: | Meng, D, Andre, P, Bateman, T.J, Berger, R, Chen, Y, Desai, K, Dewnani, S, Ellsworth, K, Feng, D, Geissler, W.M, Guo, L, Hruza, A, Jian, T, Li, H, Parker, D.L, Reichert, P, Sherer, E.C, Smith, C.J, Sonatore, L.M, Tschirret-Guth, R, Wu, J, Xu, J, Zhang, T, Campeau, L, Orr, R, Poirier, M, McCabe-Dunn, j, Araki, K, Nishimura, T, Sakurada, I, Hirabayashi, T, Wood, H.B. | | Deposit date: | 2015-10-27 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Development of a novel tricyclic class of potent and selective FIXa inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5VAT
 
 | |
5HYS
 
 | | Structure of IgE complexed with omalizumab | | Descriptor: | Epididymis luminal protein 214, Ig epsilon chain C region, SULFATE ION, ... | | Authors: | Pennington, L.F, Tarchevskaya, S.S, Sathiyamoorthy, K, Jardetzky, T.S. | | Deposit date: | 2016-02-01 | | Release date: | 2016-06-01 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of omalizumab therapy and omalizumab-mediated IgE exchange.
Nat Commun, 7, 2016
|
|
3P42
 
 | |
