6LL8
 
 | | Type II inorganic pyrophosphatase (PPase) from the psychrophilic bacterium Shewanella sp. AS-11, Mg-PNP form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, FLUORIDE ION, ... | | Authors: | Horitani, M, Kusubayashi, K, Oshima, K, Yato, A, Sugimoto, H, Watanabe, K. | | Deposit date: | 2019-12-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray Crystallography and Electron Paramagnetic Resonance Spectroscopy Reveal Active Site Rearrangement of Cold-Adapted Inorganic Pyrophosphatase.
Sci Rep, 10, 2020
|
|
6LL7
 
 | | Type II inorganic pyrophosphatase (PPase) from the psychrophilic bacterium Shewanella sp. AS-11, Mn-activated form | | Descriptor: | CALCIUM ION, Inorganic pyrophosphatase, MANGANESE (II) ION | | Authors: | Horitani, M, Kusubayashi, K, Oshima, K, Yato, A, Sugimoto, H, Watanabe, K. | | Deposit date: | 2019-12-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Crystallography and Electron Paramagnetic Resonance Spectroscopy Reveal Active Site Rearrangement of Cold-Adapted Inorganic Pyrophosphatase.
Sci Rep, 10, 2020
|
|
4YCN
 
 | | Crystal structure of the calcium pump with bound marine macrolide BLLB | | Descriptor: | (4S,5E,8S,9E,11S,13E,15E,18R)-4-hydroxy-8-methoxy-9,11-dimethyl-18-[(1Z,4E)-2-methylhexa-1,4-dien-1-yl]oxacyclooctadeca-5,9,13,15-tetraen-2-one, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Morita, M, Ogawa, H, Ohno, O, Yamori, T, Suenaga, K, Toyoshima, C. | | Deposit date: | 2015-02-20 | | Release date: | 2016-01-13 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Biselyngbyasides, cytotoxic marine macrolides, are novel and potent inhibitors of the Ca(2+) pumps with a unique mode of binding
Febs Lett., 589, 2015
|
|
4YCM
 
 | | Crystal structure of the calcium pump with bound marine macrolide BLS | | Descriptor: | (4S,5E,8S,9E,11S,13E,15E,18R)-8-methoxy-9,11-dimethyl-18-[(1Z,4E)-2-methylhexa-1,4-dien-1-yl]-2-oxooxacyclooctadeca-5,9,13,15-tetraen-4-yl 3-O-methyl-beta-D-glucopyranoside, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Morita, M, Ogawa, H, Ohno, O, Yamori, T, Suenaga, K, Toyoshima, C. | | Deposit date: | 2015-02-20 | | Release date: | 2016-01-13 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biselyngbyasides, cytotoxic marine macrolides, are novel and potent inhibitors of the Ca(2+) pumps with a unique mode of binding
Febs Lett., 589, 2015
|
|
1CKK
 
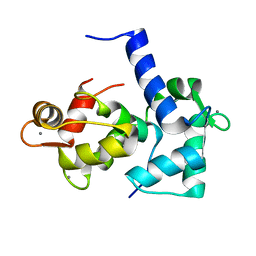 | | CALMODULIN/RAT CA2+/CALMODULIN DEPENDENT PROTEIN KINASE FRAGMENT | | Descriptor: | CALCIUM ION, Calcium/calmodulin-dependent protein kinase kinase 1, Calmodulin-1 | | Authors: | Osawa, M, Tokumitsu, H, Swindells, M.B, Kurihara, H, Orita, M, Shibanuma, T, Furuya, T, Ikura, M. | | Deposit date: | 1998-11-20 | | Release date: | 1999-09-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase.
Nat.Struct.Biol., 6, 1999
|
|
3W4K
 
 | | Crystal Structure of human DAAO in complex with coumpound 13 | | Descriptor: | 3-hydroxy-6-(2-phenylethyl)pyridazin-4(1H)-one, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3W4J
 
 | | Crystal Structure of human DAAO in complex with coumpound 12 | | Descriptor: | 3-hydroxy-5-(2-phenylethyl)pyridin-2(1H)-one, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3W4I
 
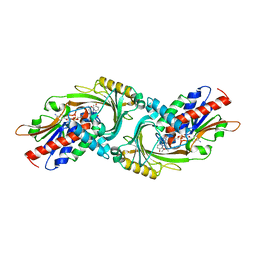 | | Crystal Structure of human DAAO in complex with coumpound 8 | | Descriptor: | D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, pyridine-2,3-diol | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
6L0W
 
 | | Structure of RLD2 BRX domain bound to LZY3 CCL motif | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, NGR2, ... | | Authors: | Hirano, Y, Futrutani, M, Nishimura, T, Taniguchi, M, Morita, M.T, Hakoshima, T. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control.
Nat Commun, 11, 2020
|
|
4U72
 
 | | Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase (A260G mutant) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-PHENYL-1H-IMIDAZOLE, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Sugimoto, H, Horitani, M, Kometani, E, Shiro, Y. | | Deposit date: | 2014-07-30 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformation and Mobility of Active Site Loop is Critical for Substrate Binding and Inhibition in Human Indoleamine 2,3-Dioxygenase
to be published
|
|
4U74
 
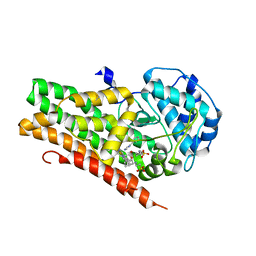 | | Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase (G262A mutant) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-PHENYL-1H-IMIDAZOLE, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Sugimoto, H, Horitani, M, Kometani, E, Shiro, Y. | | Deposit date: | 2014-07-30 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Conformation and Mobility of Active Site Loop is Critical for Substrate Binding and Inhibition in Human Indoleamine 2,3-Dioxygenase
to be published
|
|
3NGO
 
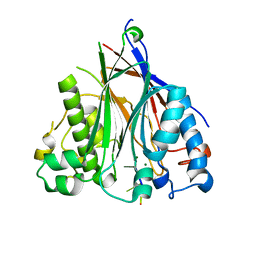 | | Crystal structure of the human CNOT6L nuclease domain in complex with poly(A) DNA | | Descriptor: | 5'-D(*AP*AP*AP*A)-3', CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-12 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
3NGQ
 
 | | Crystal structure of the human CNOT6L nuclease domain | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-13 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
3NGN
 
 | | Crystal structure of the human CNOT6L nuclease domain in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CCR4-NOT transcription complex subunit 6-like | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-12 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
3DJU
 
 | | Crystal structure of human BTG2 | | Descriptor: | Protein BTG2 | | Authors: | Yang, X, Morita, M, Wang, H, Suzuki, T, Bartlam, M, Yamamoto, T. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity
Nucleic Acids Res., 36, 2008
|
|
3DJN
 
 | | Crystal structure of mouse TIS21 | | Descriptor: | Protein BTG2 | | Authors: | Yang, X, Morita, M, Wang, H, Suzuki, T, Bartlam, M, Yamamoto, T. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity
Nucleic Acids Res., 36, 2008
|
|
6L0V
 
 | | Structure of RLD2 BRX domain bound to LZY3 CCL motif | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NGR2, ... | | Authors: | Hirano, Y, Futrutani, M, Nishimura, T, Taniguchi, M, Morita, M.T, Hakoshima, T. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.347 Å) | | Cite: | Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control.
Nat Commun, 11, 2020
|
|
7W47
 
 | | Crystal structure of the gastric proton pump complexed with tegoprazan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Potassium-transporting ATPase alpha chain 1, ... | | Authors: | Abe, K, Tanaka, S, Morita, M, Yamagishi, T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Binding of Potassium-Competitive Acid Blockers to the Gastric Proton Pump.
J.Med.Chem., 65, 2022
|
|
7W4A
 
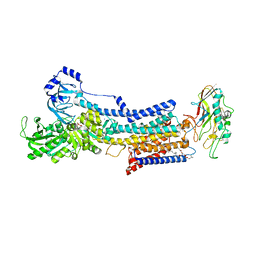 | | Cryo-EM structure of the gastric proton pump complexed with revaprazan | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Abe, K, Tanaka, S, Morita, M, Yamagishi, T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural Basis for Binding of Potassium-Competitive Acid Blockers to the Gastric Proton Pump.
J.Med.Chem., 65, 2022
|
|
