6HRB
 
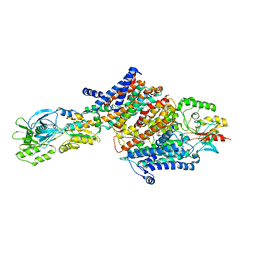 | | Cryo-EM structure of the KdpFABC complex in an E2 inward-facing state (state 2) | | Descriptor: | POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, Potassium-transporting ATPase KdpC subunit, ... | | Authors: | Stock, C, Hielkema, L, Tascon, I, Wunnicke, D, Oostergetel, G.T, Azkargorta, M, Paulino, C, Haenelt, I. | | Deposit date: | 2018-09-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of KdpFABC suggest a K+transport mechanism via two inter-subunit half-channels.
Nat Commun, 9, 2018
|
|
5MYJ
 
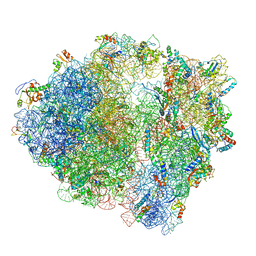 | | Structure of 70S ribosome from Lactococcus lactis | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Franken, L.E, Oostergetel, G.T, Pijning, T, Puri, P, Boekema, E.J, Poolman, B, Guskov, A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy.
Nat Commun, 8, 2017
|
|
6GCT
 
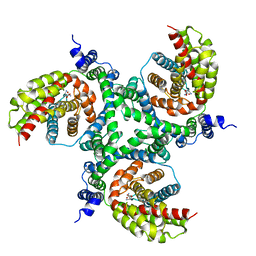 | | cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Garaeva, A.A, Oostergetel, G.T, Gati, C, Guskov, A, Paulino, C, Slotboom, D.J. | | Deposit date: | 2018-04-19 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM structure of the human neutral amino acid transporter ASCT2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5MDX
 
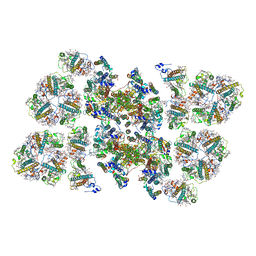 | | Cryo-EM structure of the PSII supercomplex from Arabidopsis thaliana | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL B, Chlorophyll a-b binding protein 1, ... | | Authors: | van Bezouwen, L.S, Caffarri, S, Kale, R.S, Kouril, R, Thunnissen, A.M.W.H, Oostergetel, G.T, Boekema, E.J. | | Deposit date: | 2016-11-13 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Subunit and chlorophyll organization of the plant photosystem II supercomplex.
Nat Plants, 3, 2017
|
|
6HRA
 
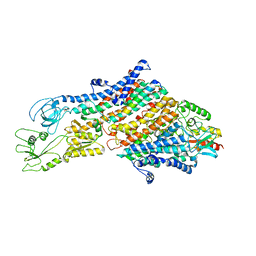 | | Cryo-EM structure of the KdpFABC complex in an E1 outward-facing state (state 1) | | Descriptor: | POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, Potassium-transporting ATPase KdpC subunit, ... | | Authors: | Stock, C, Hielkema, L, Tascon, I, Wunnicke, D, Oostergetel, G.T, Azkargorta, M, Paulino, C, Haenelt, I. | | Deposit date: | 2018-09-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of KdpFABC suggest a K+transport mechanism via two inter-subunit half-channels.
Nat Commun, 9, 2018
|
|
6QMB
 
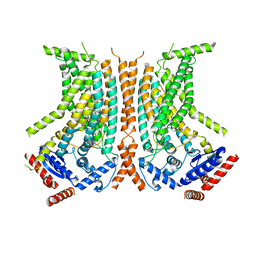 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (closed state) | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QPB
 
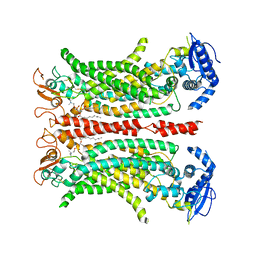 | | Cryo-EM structure of calcium-free mTMEM16F lipid scramblase in digitonin | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6 | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
6QM9
 
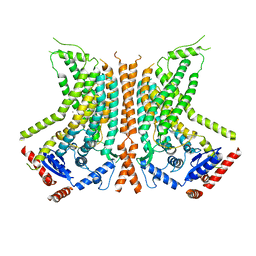 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (open state) | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QP6
 
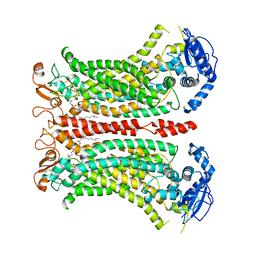 | | Cryo-EM structure of calcium-bound mTMEM16F lipid scramblase in digitonin | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
6QMA
 
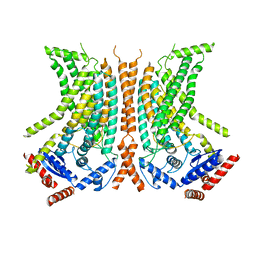 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (intermediate state) | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QPI
 
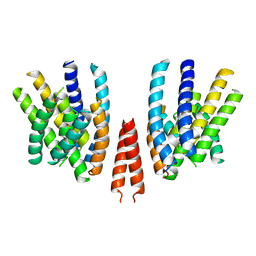 | | Cryo-EM structure of calcium-free mTMEM16F lipid scramblase in nanodisc | | Descriptor: | Anoctamin-6 | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
6QM5
 
 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in DDM | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QPC
 
 | | Cryo-EM structure of calcium-bound mTMEM16F lipid scramblase in nanodisc | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-13 | | Release date: | 2019-03-06 | | Last modified: | 2019-04-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
6QM4
 
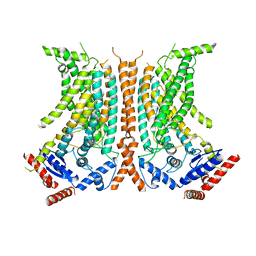 | | Cryo-EM structure of calcium-free nhTMEM16 lipid scramblase in nanodisc | | Descriptor: | Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QM6
 
 | | Cryo-EM structure of calcium-free nhTMEM16 lipid scramblase in DDM | | Descriptor: | Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
