3ZNF
 
 | |
3GAT
 
 | | SOLUTION NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF CHICKEN GATA-1 BOUND TO DNA, 34 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*TP*CP*TP*GP*CP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*AP*GP*AP*TP*AP*AP*AP*CP*AP*TP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | Authors: | Clore, G.M, Tjandra, N, Starich, M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution.
Nat.Struct.Biol., 4, 1997
|
|
4ZNF
 
 | |
8DJH
 
 | | Ternary complex of SUMO1 with a phosphomimetic SIM of PML and zinc | | Descriptor: | PML 4SD, Small ubiquitin-related modifier 1, ZINC ION | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Bourdeau, V, Gagnon, C, Igelmann, S, Sakaguchi, K, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Zinc controls PML nuclear body formation through regulation of a paralog specific auto-inhibition in SUMO1.
Nucleic Acids Res., 50, 2022
|
|
8DJI
 
 | | Ternary complex of SUMO1 with the SIM of PML and zinc | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1, ZINC ION | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Bourdeau, V, Gagnon, C, Igelmann, S, Sakaguchi, K, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Zinc controls PML nuclear body formation through regulation of a paralog specific auto-inhibition in SUMO1.
Nucleic Acids Res., 50, 2022
|
|
2GAT
 
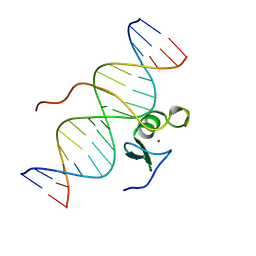 | | SOLUTION STRUCTURE OF THE C-TERMINAL DOMAIN OF CHICKEN GATA-1 BOUND TO DNA, NMR, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*TP*CP*TP*GP*CP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*AP*GP*AP*TP*AP*AP*AP*CP*AP*TP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | Authors: | Clore, G.M, Tjandra, N, Starich, M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution.
Nat.Struct.Biol., 4, 1997
|
|
3F2F
 
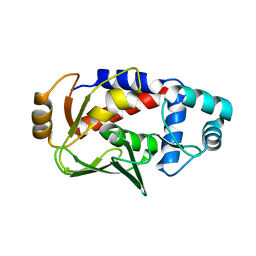 | | Crystal structure of the mercury-bound form of MerB, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
1GAT
 
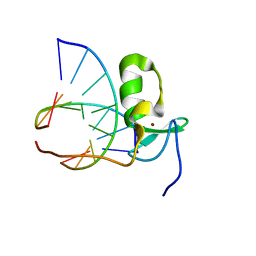 | | SOLUTION STRUCTURE OF THE SPECIFIC DNA COMPLEX OF THE ZINC CONTAINING DNA BINDING DOMAIN OF THE ERYTHROID TRANSCRIPTION FACTOR GATA-1 BY MULTIDIMENSIONAL NMR | | Descriptor: | DNA (5'-D(P*AP*GP*AP*TP*AP*AP*AP*C)3'), DNA (5'-D(P*GP*TP*TP*TP*AP*TP*CP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | Authors: | Clore, G.M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1993-06-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1.
Science, 261, 1993
|
|
1GAU
 
 | |
2GS0
 
 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and the activation domain of p53 | | Descriptor: | Cellular tumor antigen p53, RNA polymerase II transcription factor B subunit 1 | | Authors: | Di Lello, P, Jones, T.N, Nguyen, B.D, Legault, P, Omichinski, J.G. | | Deposit date: | 2006-04-25 | | Release date: | 2006-10-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53.
Mol.Cell, 22, 2006
|
|
1SAK
 
 | |
1SAE
 
 | |
1SAF
 
 | |
1SAL
 
 | |
1Y5O
 
 | | NMR structure of the amino-terminal domain from the Tfb1 subunit of yeast TFIIH | | Descriptor: | RNA polymerase II transcription factor B 73 kDa subunit | | Authors: | Di Lello, P, Nguyen, B.D, Jones, T.N, Potempa, K, Kobor, M.S, Legault, P, Omichinski, J.G. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Amino-Terminal Domain from the Tfb1 Subunit of TFIIH and
Characterization of Its Phosphoinositide and VP16 Binding Sites
Biochemistry, 44, 2005
|
|
1YUI
 
 | | SOLUTION NMR STRUCTURE OF THE GAGA FACTOR/DNA COMPLEX, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*AP*GP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*TP*CP*TP*CP*GP*GP*C)-3'), GAGA-FACTOR, ... | | Authors: | Clore, G.M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1996-12-31 | | Release date: | 1997-12-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode.
Nat.Struct.Biol., 4, 1997
|
|
1YUJ
 
 | | SOLUTION NMR STRUCTURE OF THE GAGA FACTOR/DNA COMPLEX, 50 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*AP*GP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*TP*CP*TP*CP*GP*GP*C)-3'), GAGA-FACTOR, ... | | Authors: | Clore, G.M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1996-12-31 | | Release date: | 1997-12-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode.
Nat.Struct.Biol., 4, 1997
|
|
1BBO
 
 | |
1DL6
 
 | | SOLUTION STRUCTURE OF HUMAN TFIIB N-TERMINAL DOMAIN | | Descriptor: | TRANSCRIPTION FACTOR II B (TFIIB), ZINC ION | | Authors: | Chen, H.-T, Legault, P, Glushka, J, Omichinski, J.G, Scott, R.A. | | Deposit date: | 1999-12-08 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a (Cys3His) zinc ribbon, a ubiquitous motif in archaeal and eucaryal transcription.
Protein Sci., 9, 2000
|
|
8T2N
 
 | | Crystal structure of GABARAP in complex with the LIR of NSs3 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, Non-structural protein S | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
8T2L
 
 | | Crystal structure of LC3A in complex with the LIR of NSs4 | | Descriptor: | Non-structural protein S,Microtubule-associated proteins 1A/1B light chain 3A chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
8T2M
 
 | | Crystal structure of GABARAP in complex with the LIR of NSs4 | | Descriptor: | Non-structural protein S,Gamma-aminobutyric acid receptor-associated protein chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
5C0T
 
 | | Crystal structure of the mercury-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
5DSF
 
 | | Crystal structure of the mercury-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-09-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
5C17
 
 | | Crystal structure of the mercury-bound form of MerB2 | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-13 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
