2F87
 
 | | Solution structure of a GAAG tetraloop in SRP RNA from Pyrococcus furiosus | | Descriptor: | SRP RNA | | Authors: | Okada, K, Takahashi, M, Sakamoto, T, Nakamura, K, Kanai, A, Kawai, G | | Deposit date: | 2005-12-02 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a GAAG tetraloop in helix 6 of SRP RNA from Pyrococcus furiosus
Nucleosides Nucleotides Nucleic Acids, 25, 2006
|
|
5VAD
 
 | | Crystal structure of human Prolyl-tRNA synthetase (PRS) in complex with inhibitor | | Descriptor: | 3-[(cyclohexanecarbonyl)amino]-N-(2,3-dihydro-1H-inden-2-yl)pyrazine-2-carboxamide, Bifunctional glutamate/proline--tRNA ligase, PROLINE, ... | | Authors: | Okada, K, Skene, R.J. | | Deposit date: | 2017-03-24 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery of a novel prolyl-tRNA synthetase inhibitor and elucidation of its binding mode to the ATP site in complex with l-proline.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
2FYH
 
 | | Solution structure of the 2'-5' RNA ligase-like protein from Pyrococcus furiosus | | Descriptor: | putative integral membrane transport protein | | Authors: | Okada, K, Matsuda, T, Sakamoto, T, Muto, Y, Yokoyama, S, Kanai, A, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Characterization of a heat-stable enzyme possessing GTP-dependent RNA ligase activity from a hyperthermophilic archaeon, Pyrococcus furiosus
Rna, 15, 2009
|
|
6JZJ
 
 | | Structure of FimA type-2 (FimA2) prepilin of the type V major fimbrium | | Descriptor: | Major fimbrium subunit FimA type-2, SULFATE ION | | Authors: | Okada, K, Shoji, M, Nakayam, K, Imada, K. | | Deposit date: | 2019-05-02 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange.
Nat Microbiol, 5, 2020
|
|
6JZK
 
 | | Structure of FimA type-1 (FimA1) prepilin of the type V major fimbrium | | Descriptor: | GLYCEROL, Major fimbrium subunit FimA type-1, S,R MESO-TARTARIC ACID, ... | | Authors: | Okada, K, Shoji, M, Nakayam, K, Imada, K. | | Deposit date: | 2019-05-02 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange.
Nat Microbiol, 5, 2020
|
|
1A3G
 
 | | BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE FROM ESCHERICHIA COLI | | Descriptor: | BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okada, K, Hirotsu, K, Sato, M, Hayashi, H, Kagamiyama, H. | | Deposit date: | 1998-01-21 | | Release date: | 1998-05-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of Escherichia coli branched-chain amino acid aminotransferase at 2.5 A resolution.
J.Biochem.(Tokyo), 121, 1997
|
|
1I1L
 
 | | CRYSTAL STRUCTURE OF ESCHELICHIA COLI BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE. | | Descriptor: | 2-METHYLLEUCINE, BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okada, K, Hirotsu, K, Hayashi, H, Kagamiyama, H. | | Deposit date: | 2001-02-02 | | Release date: | 2001-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Escherichia coli branched-chain amino acid aminotransferase and its complexes with 4-methylvalerate and 2-methylleucine: induced fit and substrate recognition of the enzyme.
Biochemistry, 40, 2001
|
|
1I1M
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE. | | Descriptor: | 4-METHYL VALERIC ACID, BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okada, K, Hirotsu, K, Hayashi, H, Kagamiyama, H. | | Deposit date: | 2001-02-02 | | Release date: | 2001-07-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Escherichia coli branched-chain amino acid aminotransferase and its complexes with 4-methylvalerate and 2-methylleucine: induced fit and substrate recognition of the enzyme.
Biochemistry, 40, 2001
|
|
3WX9
 
 | | Crystal structure of Pyrococcus horikoshii kynurenine aminotransferase in complex with PMP, GLA, 4AD, 2OG, GLU and KYA | | Descriptor: | (2E)-pent-2-enedioic acid, 2-OXOGLUTARIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Okada, K, Angkawidjaja, C, Koga, Y, Kanaya, S. | | Deposit date: | 2014-07-28 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of Pyrococcus horikoshii kynurenine aminotransferase in complex with PMP, GLA, 4AD, 2OG, GLU and KYA
To be Published
|
|
1I1K
 
 | | CRYSTAL STRUCTURE OF ESCHELICHIA COLI BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE. | | Descriptor: | BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okada, K, Hirotsu, K, Hayashi, H, Kagamiyama, H. | | Deposit date: | 2001-02-02 | | Release date: | 2001-07-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Escherichia coli branched-chain amino acid aminotransferase and its complexes with 4-methylvalerate and 2-methylleucine: induced fit and substrate recognition of the enzyme.
Biochemistry, 40, 2001
|
|
3AOV
 
 | | Crystal structure of Pyrococcus horikoshii kynurenine aminotransferase in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative uncharacterized protein PH0207 | | Authors: | Okada, K, Angkawidjaja, C, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2010-10-07 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Characterization of kynurenine aminotransferase from hyperthermophilic archaeon: enzymatic activity for conversion to kynurenic acid is allosterically regulated by alpha-ketoglutaric acid cooperatively with kynurenine
To be Published
|
|
3ATH
 
 | | Crystal structure of Pyrococcus horikoshii kynurenine aminotransferase in complex with four AKGs as substrates and allosteric effectors | | Descriptor: | 2-OXOGLUTARIC ACID, PYRIDOXAL-5'-PHOSPHATE, Putative uncharacterized protein PH0207 | | Authors: | Okada, K, Angkawidjaja, C, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2011-01-05 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Characterization of kynurenine aminotransferase from hyperthermophilic archaeon: enzymatic activity for conversion to kynurenic acid is allosterically regulated by alpha-ketoglutaric acid cooperatively with kynurenine
To be Published
|
|
3AV7
 
 | | Crystal structure of Pyrococcus horikoshii kynurenine aminotransferase in complex with PMP, KYN as substrates and KYA as products | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-hydroxyquinoline-2-carboxylic acid, ... | | Authors: | Okada, K, Angkawidjaja, C, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2011-02-23 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural insight into kynurenic acid excretion mechanisms of kynurenine aminotransferase in the hyperthermophilic archaeon Pyrococcus horikoshii
To be Published
|
|
3AOW
 
 | | Crystal structure of Pyrococcus horikoshii kynurenine aminotransferase in complex with AKG | | Descriptor: | 2-OXOGLUTARIC ACID, PYRIDOXAL-5'-PHOSPHATE, Putative uncharacterized protein PH0207 | | Authors: | Okada, K, Angkawidjaja, C, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2010-10-07 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Characterization of kynurenine aminotransferase from hyperthermophilic archaeon: enzymatic activity for conversion to kynurenic acid is allosterically regulated by alpha-ketoglutaric acid cooperatively with kynurenine
To be Published
|
|
2Z04
 
 | | Crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit from Aquifex aeolicus | | Descriptor: | Phosphoribosylaminoimidazole carboxylase ATPase subunit, SULFATE ION | | Authors: | Okada, K, Tamura, S, Baba, S, Kanagawa, M, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit from Aquifex aeolicus
To be Published
|
|
3AW8
 
 | | Crystal structure of N5-carboxyaminoimidazole ribonucleotide synthetase from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Phosphoribosylaminoimidazole carboxylase, ... | | Authors: | Okada, K, Tsunoda, S, Taka, H, Baba, S, Kanagawa, M, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-03-15 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of N5-carboxyaminoimidazole ribonucleotide synthetase, PurK, from thermophilic bacteria
To be Published
|
|
1QQ2
 
 | | CRYSTAL STRUCTURE OF A MAMMALIAN 2-CYS PEROXIREDOXIN, HBP23. | | Descriptor: | CHLORIDE ION, THIOREDOXIN PEROXIDASE 2 | | Authors: | Hirotsu, S, Abe, Y, Okada, K, Nagahara, N, Hori, H, Nishino, T, Hakoshima, T. | | Deposit date: | 1999-06-10 | | Release date: | 1999-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a multifunctional 2-Cys peroxiredoxin heme-binding protein 23 kDa/proliferation-associated gene product.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6M9L
 
 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridine-2-one based p38 MAP Kinase Inhibitors by scaffold hopping - compound 10 | | Descriptor: | 3-benzyl-6-[(2,4-difluorophenyl)amino]-1,3-dihydro-2H-imidazo[4,5-b]pyridin-2-one, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Okada, K. | | Deposit date: | 2018-08-23 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 1.
Chemmedchem, 14, 2019
|
|
6M95
 
 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridine-2-one based p38 MAP Kinase Inhibitors by scaffold hopping: compound 1 | | Descriptor: | (4-benzylpiperidin-1-yl)[2-methoxy-4-(methylsulfanyl)phenyl]methanone, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Okada, K. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 1.
Chemmedchem, 14, 2019
|
|
4PHU
 
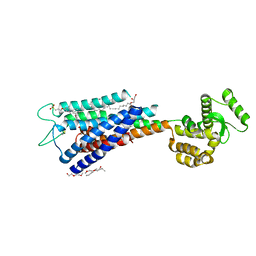 | | Crystal structure of Human GPR40 bound to allosteric agonist TAK-875 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DIMETHYL SULFOXIDE, Free fatty acid receptor 1,Lysozyme, ... | | Authors: | Srivastava, A, Yano, J.K, Hirozane, Y, Kefala, G, Snell, G, Lane, W, Gruswitz, F, Ivetac, A, Aertgeerts, K, Nguyen, J, Jennings, A, Okada, K. | | Deposit date: | 2014-05-07 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875.
Nature, 513, 2014
|
|
8WO8
 
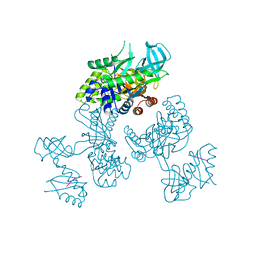 | | Crystal Structure of an RNA-binding protein, FAU-1, from Pyrococcus furiosus | | Descriptor: | Probable ribonuclease FAU-1, RNA (5'-R(P*AP*UP*A)-3') | | Authors: | Kawai, G, Okada, K, Baba, S, Sato, A, Sakamoto, T, Kanai, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Homo-trimeric structure of the ribonuclease for rRNA processing, FAU-1, from Pyrococcus furiosus.
J.Biochem., 175, 2024
|
|
3J67
 
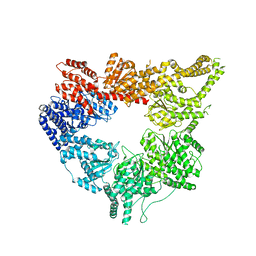 | | Structural mechanism of the dynein powerstroke (post-powerstroke state) | | Descriptor: | Dynein motor domain | | Authors: | Lin, J, Okada, K, Raytchev, M, Smith, M.C, Nicastro, D. | | Deposit date: | 2013-12-22 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (34 Å) | | Cite: | Structural mechanism of the dynein power stroke.
Nat.Cell Biol., 16, 2014
|
|
3J68
 
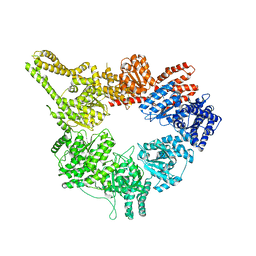 | | Structural mechanism of the dynein powerstroke (pre-powerstroke state) | | Descriptor: | Dynein motor domain | | Authors: | Lin, J, Okada, K, Raytchev, M, Smith, M.C, Nicastro, D. | | Deposit date: | 2013-12-23 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Structural mechanism of the dynein power stroke.
Nat.Cell Biol., 16, 2014
|
|
1WPL
 
 | | Crystal structure of the inhibitory form of rat GTP cyclohydrolase I/GFRP complex | | Descriptor: | 7,8-DIHYDROBIOPTERIN, GTP cyclohydrolase I, GTP cyclohydrolase I feedback regulatory protein, ... | | Authors: | Maita, N, Hatakeyama, K, Okada, K, Hakoshima, T. | | Deposit date: | 2004-09-08 | | Release date: | 2004-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of biopterin-induced inhibition of GTP cyclohydrolase I by GFRP, its feedback regulatory protein
J.Biol.Chem., 279, 2004
|
|
2HI7
 
 | | Crystal structure of DsbA-DsbB-ubiquinone complex | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein dsbA, UBIQUINONE-1, ... | | Authors: | Inaba, K, Murakami, S, Suzuki, M, Nakagawa, A, Yamashita, E, Okada, K, Ito, K. | | Deposit date: | 2006-06-29 | | Release date: | 2006-12-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of the DsbB-DsbA Complex Reveals a Mechanism of Disulfide Bond Generation
Cell(Cambridge,Mass.), 127, 2006
|
|
