7F1I
 
 | | Designed enzyme RA61 M48K/I72D mutant: form II | | 分子名称: | Engineered Retroaldolase | | 著者 | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | 登録日 | 2021-06-09 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1H
 
 | | Designed enzyme RA61 M48K/I72D mutant: form I | | 分子名称: | Engineered Retroaldolase, FORMIC ACID, GLYCEROL | | 著者 | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | 登録日 | 2021-06-09 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.14 Å) | | 主引用文献 | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1J
 
 | | Designed enzyme RA61 M48K/I72D mutant: form III | | 分子名称: | Engineered Retroaldolase | | 著者 | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | 登録日 | 2021-06-09 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1K
 
 | | Designed enzyme RA61 M48K/I72D mutant: form IV | | 分子名称: | Engineered Retroaldolase | | 著者 | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | 登録日 | 2021-06-09 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1L
 
 | | Designed enzyme RA61 M48K/I72D mutant: form V | | 分子名称: | CHLORIDE ION, Engineered Retroaldolase, IMIDAZOLE | | 著者 | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | 登録日 | 2021-06-09 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
3X3Y
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by histamine | | 分子名称: | COPPER (II) ION, GLYCEROL, POTASSIUM ION, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.499 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X42
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis in the presence of sodium bromide | | 分子名称: | BROMIDE ION, COPPER (II) ION, GLYCEROL, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.875 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X41
 
 | | Copper amine oxidase from Arthrobacter globiformis: Product Schiff-base form produced by anaerobic reduction in the presence of sodium bromide | | 分子名称: | BROMIDE ION, COPPER (II) ION, GLYCEROL, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X3X
 
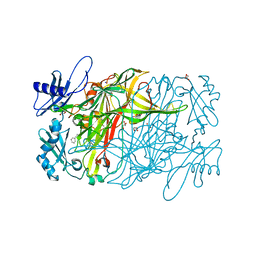 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine | | 分子名称: | 2-PHENYL-ETHANOL, COPPER (II) ION, GLYCEROL, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X3Z
 
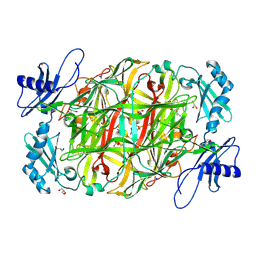 | | Copper amine oxidase from Arthrobacter globiformis: Aminoresorcinol form produced by anaerobic reduction with ethylamine hydrochloride | | 分子名称: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X40
 
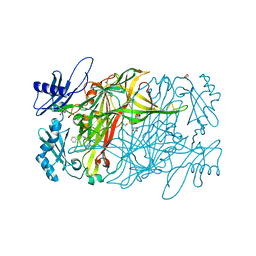 | | Copper amine oxidase from Arthrobacter globiformis: Product Schiff-base form produced by anaerobic reduction in the presence of sodium chloride | | 分子名称: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
5B1O
 
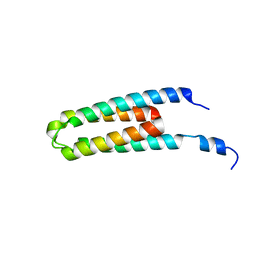 | | DHp domain structure of EnvZ P248A mutant | | 分子名称: | Osmolarity sensor protein EnvZ | | 著者 | Okajima, T, Eguchi, Y, Tochio, N, Inukai, Y, Shimizu, R, Ueda, S, Shinya, S, Kigawa, T, Fukamizo, T, Igarashi, M, Utsumi, R. | | 登録日 | 2015-12-09 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases
J. Antibiot., 70, 2017
|
|
1MQA
 
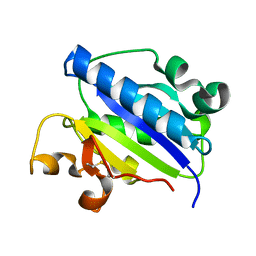 | | Crystal structure of high affinity alphaL I domain in the absence of ligand or metal | | 分子名称: | Integrin alpha-L | | 著者 | Shimaoka, T, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, Zhang, R, Takagi, J, Wang, J.-H, Springer, T.A. | | 登録日 | 2002-09-15 | | 公開日 | 2003-01-14 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
5B1N
 
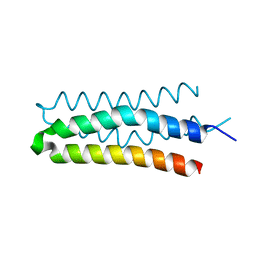 | | DHp domain structure of EnvZ from Escherichia coli | | 分子名称: | Osmolarity sensor protein EnvZ | | 著者 | Okajima, T, Eguchi, Y, Tochio, N, Inukai, Y, Shimizu, R, Ueda, S, Shinya, S, Kigawa, T, Fukamizo, T, Igarashi, M, Utsumi, R. | | 登録日 | 2015-12-09 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases
J. Antibiot., 70, 2017
|
|
7WEW
 
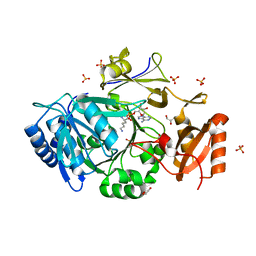 | | Structure of adenylation domain of epsilon-poly-L-lysine synthase | | 分子名称: | ADENOSINE-5'-[LYSYL-PHOSPHATE], Epsilon-poly-L-lysine synthase, GLYCEROL, ... | | 著者 | Okamoto, T, Yamanaka, K, Hamano, Y, Nagano, S, Hino, T. | | 登録日 | 2021-12-24 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the adenylation domain from an epsilon-poly-l-lysine synthetase provides molecular mechanism for substrate specificity
Biochem.Biophys.Res.Commun., 596, 2022
|
|
5Z98
 
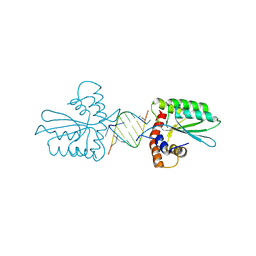 | | Crystal Structure of the Primate APOBEC3H Dimer mediated by RNA Duplex | | 分子名称: | Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like protein 3H, RNA (5'-R(*AP*UP*AP*CP*CP*CP*GP*GP*CP*A)-3'), RNA (5'-R(P*CP*UP*GP*CP*CP*GP*GP*GP*UP*A)-3'), ... | | 著者 | Matsuoka, T, Nagae, T, Ode, H, Watanabe, N, Iwatani, Y. | | 登録日 | 2018-02-02 | | 公開日 | 2018-08-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of chimpanzee APOBEC3H dimerization stabilized by double-stranded RNA.
Nucleic Acids Res., 46, 2018
|
|
1L9H
 
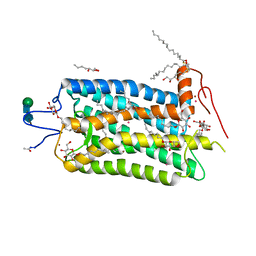 | | Crystal structure of bovine rhodopsin at 2.6 angstroms RESOLUTION | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | 著者 | Okada, T, Fujiyoshi, Y, Silow, M, Navarro, J, Landau, E.M, Shichida, Y. | | 登録日 | 2002-03-23 | | 公開日 | 2002-05-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1F88
 
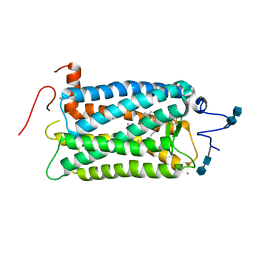 | | CRYSTAL STRUCTURE OF BOVINE RHODOPSIN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MERCURY (II) ION, RETINAL, ... | | 著者 | Okada, T, Palczewski, K, Stenkamp, R.E, Miyano, M. | | 登録日 | 2000-06-29 | | 公開日 | 2000-08-04 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of rhodopsin: A G protein-coupled receptor.
Science, 289, 2000
|
|
2ZJV
 
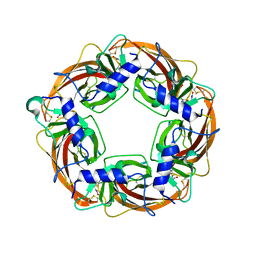 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein (Ls-AChBP) Complexed with Clothianidin | | 分子名称: | 1-[(2-chloro-1,3-thiazol-5-yl)methyl]-3-methyl-2-nitroguanidine, Acetylcholine-binding protein | | 著者 | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Morimoto, T, Matsuda, K. | | 登録日 | 2008-03-10 | | 公開日 | 2008-04-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structures of Lymnaea stagnalis AChBP in complex with neonicotinoid insecticides imidacloprid and clothianidin
Invert.Neurosci., 8, 2008
|
|
7YRO
 
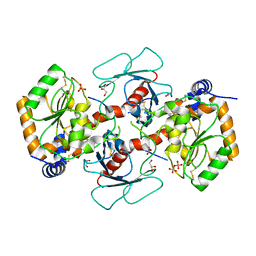 | | Crystal structure of mango fucosyltransferase 13 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, Fucosyltransferase, ... | | 著者 | Okada, T, Teramoto, T, Ihara, H, Ikeda, Y, Kakuta, Y. | | 登録日 | 2022-08-10 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Crystal structure of mango alpha 1,3/ alpha 1,4-fucosyltransferase elucidates unique elements that regulate Lewis A-dominant oligosaccharide assembly.
Glycobiology, 34, 2024
|
|
2ZJU
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein (Ls-AChBP) Complexed with Imidacloprid | | 分子名称: | (2E)-1-[(6-chloropyridin-3-yl)methyl]-N-nitroimidazolidin-2-imine, Acetylcholine-binding protein | | 著者 | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Morimoto, T, Matsuda, K. | | 登録日 | 2008-03-10 | | 公開日 | 2008-04-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Crystal structures of Lymnaea stagnalis AChBP in complex with neonicotinoid insecticides imidacloprid and clothianidin
Invert.Neurosci., 8, 2008
|
|
2ZTK
 
 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with homocitrate | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, COPPER (II) ION, Homocitrate synthase | | 著者 | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | 登録日 | 2008-10-06 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from Thermus thermophilus
J.Biol.Chem., 285, 2010
|
|
2ZTJ
 
 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, COPPER (II) ION, Homocitrate synthase | | 著者 | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | 登録日 | 2008-10-06 | | 公開日 | 2009-10-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from Thermus thermophilus
J.Biol.Chem., 285, 2010
|
|
2ZYF
 
 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with magnesuim ion and alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, Homocitrate synthase, MAGNESIUM ION | | 著者 | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | 登録日 | 2009-01-20 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from thermus thermophilus
J.Biol.Chem., 2009
|
|
3A9I
 
 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with Lys | | 分子名称: | COBALT (II) ION, Homocitrate synthase, LYSINE | | 著者 | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | 登録日 | 2009-10-28 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from Thermus thermophilus
J.Biol.Chem., 285, 2010
|
|
