1DIO
 
 | | DIOL DEHYDRATASE-CYANOCOBALAMIN COMPLEX FROM KLEBSIELLA OXYTOCA | | 分子名称: | COBALAMIN, POTASSIUM ION, PROTEIN (DIOL DEHYDRATASE), ... | | 著者 | Shibata, N, Masuda, J, Tobimatsu, T, Toraya, T, Suto, K, Morimoto, Y, Yasuoka, N. | | 登録日 | 1999-01-27 | | 公開日 | 2000-01-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A new mode of B12 binding and the direct participation of a potassium ion in enzyme catalysis: X-ray structure of diol dehydratase.
Structure Fold.Des., 7, 1999
|
|
1EEX
 
 | | CRYSTAL STRUCTURE OF THE DIOL DEHYDRATASE-ADENINYLPENTYLCOBALAMIN COMPLEX FROM KLEBSIELLA OXYTOCA | | 分子名称: | CO-(ADENIN-9-YL-PENTYL)-COBALAMIN, POTASSIUM ION, PROPANEDIOL DEHYDRATASE, ... | | 著者 | Shibata, N, Masuda, J, Toraya, T, Morimoto, Y, Yasuoka, N. | | 登録日 | 2000-02-04 | | 公開日 | 2001-02-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | How a protein generates a catalytic radical from coenzyme B(12): X-ray structure of a diol-dehydratase-adeninylpentylcobalamin complex.
Structure Fold.Des., 8, 2000
|
|
1EGV
 
 | | CRYSTAL STRUCTURE OF THE DIOL DEHYDRATASE-ADENINYLPENTYLCOBALAMIN COMPLEX FROM KLEBSELLA OXYTOCA UNDER THE ILLUMINATED CONDITION. | | 分子名称: | CO-(ADENIN-9-YL-PENTYL)-COBALAMIN, POTASSIUM ION, PROPANEDIOL DEHYDRATASE, ... | | 著者 | Masuda, J, Shibata, N, Toraya, T, Morimoto, Y, Yasuoka, N. | | 登録日 | 2000-02-17 | | 公開日 | 2001-02-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | How a protein generates a catalytic radical from coenzyme B(12): X-ray structure of a diol-dehydratase-adeninylpentylcobalamin complex.
Structure Fold.Des., 8, 2000
|
|
1EGM
 
 | | CRYSTAL STRUCTURE OF DIOL DEHYDRATASE-CYANOCOBALAMIN COMPLEX AT 100K. | | 分子名称: | CYANOCOBALAMIN, POTASSIUM ION, PROPANEDIOL DEHYDRATASE, ... | | 著者 | Masuda, J, Shibata, N, Toraya, T, Morimoto, Y, Yasuoka, N. | | 登録日 | 2000-02-15 | | 公開日 | 2000-09-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | How a protein generates a catalytic radical from coenzyme B(12): X-ray structure of a diol-dehydratase-adeninylpentylcobalamin complex.
Structure Fold.Des., 8, 2000
|
|
5XXX
 
 | | GMPCPP-microtubule complexed with nucleotide-free KIF5C | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | 著者 | Morikawa, M, Shigematsu, H, Nitta, R, Hirokawa, N. | | 登録日 | 2017-07-05 | | 公開日 | 2018-10-10 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (6.43 Å) | | 主引用文献 | Kinesin-binding-triggered conformation switching of microtubules contributes to polarized transport
J. Cell Biol., 217, 2018
|
|
4LGW
 
 | | Crystal structure of Escherichia coli SdiA in the space group P6522 | | 分子名称: | GLYCEROL, Regulatory protein SdiA | | 著者 | Kim, T, Duong, T, Wu, C.A, Choi, J, Lan, N, Kang, S.W, Lokanath, N.K, Shin, D, Hwang, H.Y, Kim, K.K. | | 登録日 | 2013-06-28 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5XXV
 
 | | GDP-microtubule complexed with KIF5C in AMPPNP state | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Morikawa, M, Shigematsu, H, Nitta, R, Hirokawa, N. | | 登録日 | 2017-07-05 | | 公開日 | 2018-10-10 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (6.46 Å) | | 主引用文献 | Kinesin-binding-triggered conformation switching of microtubules contributes to polarized transport
J. Cell Biol., 217, 2018
|
|
4LFU
 
 | | Crystal structure of Escherichia coli SdiA in the space group C2 | | 分子名称: | CHLORIDE ION, Regulatory protein SdiA, TETRAETHYLENE GLYCOL | | 著者 | Kim, T, Duong, T, Wu, C.A, Choi, J, Lan, N, Kang, S.W, Lokanath, N.K, Shin, D, Hwang, H.Y, Kim, K.K. | | 登録日 | 2013-06-27 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JP3
 
 | | Crystal Structure of TT0495 protein from Thermus thermophilus HB8 | | 分子名称: | 2-deoxy-D-gluconate 3-dehydrogenase, CITRIC ACID | | 著者 | Pampa, K.J, Lokanath, N.K, Kunishima, N, Ravishnkar Rai, V. | | 登録日 | 2013-03-19 | | 公開日 | 2014-03-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The first crystal structure of NAD-dependent 3-dehydro-2-deoxy-D-gluconate dehydrogenase from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1FLM
 
 | | DIMER OF FMN-BINDING PROTEIN FROM DESULFOVIBRIO VULGARIS (MIYAZAKI F) | | 分子名称: | FLAVIN MONONUCLEOTIDE, PROTEIN (FMN-BINDING PROTEIN) | | 著者 | Suto, K, Kawagoe, K, Shibata, N, Morimoto, K, Higuchi, Y, Kitamura, M, Nakaya, T, Yasuoka, N. | | 登録日 | 1999-03-10 | | 公開日 | 2000-03-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | How do the x-ray structure and the NMR structure of FMN-binding protein differ?
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1EQQ
 
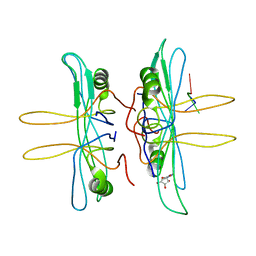 | | SINGLE STRANDED DNA BINDING PROTEIN AND SSDNA COMPLEX | | 分子名称: | 5'-R(*(5MU)P*(5MU)P*(5MU))-3', SINGLE STRANDED DNA BINDING PROTEIN | | 著者 | Matsumoto, T, Morimoto, Y, Shibata, N, Yasuoka, N, Shimamoto, N. | | 登録日 | 2000-04-06 | | 公開日 | 2003-09-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Roles of functional loops and the C-terminal segment of a single-stranded DNA binding protein elucidated by X-Ray structure analysis
J.Biochem.(Tokyo), 127, 2000
|
|
1H2A
 
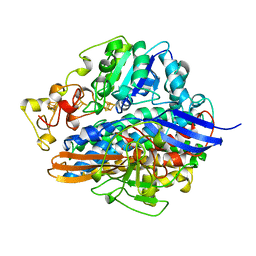 | |
1WYT
 
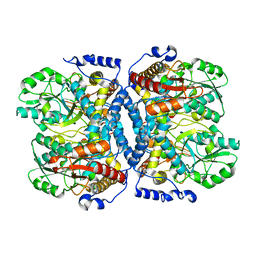 | | Crystal structure of glycine decarboxylase (P-protein) of the glycine cleavage system, in apo form | | 分子名称: | glycine dehydrogenase (decarboxylating) subunit 1, glycine dehydrogenase subunit 2 (P-protein) | | 著者 | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-02-17 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of P-protein of the glycine cleavage system: implications for nonketotic hyperglycinemia
Embo J., 24, 2005
|
|
7C7G
 
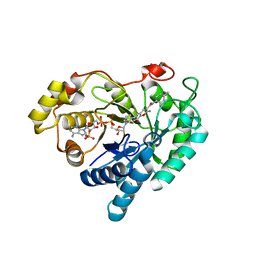 | |
7VJZ
 
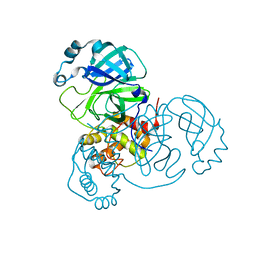 | |
1WYV
 
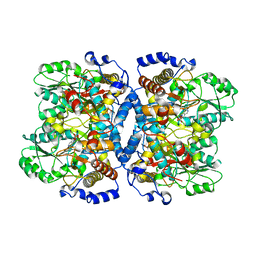 | | Crystal structure of glycine decarboxylase (P-protein) of the glycine cleavage system, in inhibitor-bound form | | 分子名称: | (AMINOOXY)ACETIC ACID, PYRIDOXAL-5'-PHOSPHATE, glycine dehydrogenase (decarboxylating) subunit 1, ... | | 著者 | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-02-17 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of P-protein of the glycine cleavage system: implications for nonketotic hyperglycinemia
Embo J., 24, 2005
|
|
2RVO
 
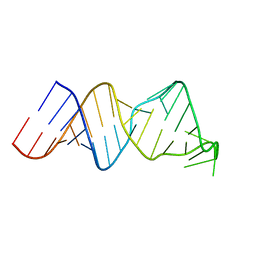 | | Solution structure of a reverse transcriptase recognition site of a LINE RNA from zebrafish | | 分子名称: | RNA (34-MER) | | 著者 | Otsu, M, Norose, N, Arai, N, Terao, R, Kajikawa, M, Okada, N, Kawai, G. | | 登録日 | 2016-02-03 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a reverse transcriptase recognition site of a LINE RNA from zebrafish.
J. Biochem., 162, 2017
|
|
1WYU
 
 | | Crystal structure of glycine decarboxylase (P-protein) of the glycine cleavage system, in holo form | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, glycine dehydrogenase (decarboxylating) subunit 1, glycine dehydrogenase subunit 2 (P-protein) | | 著者 | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-02-17 | | 公開日 | 2005-04-05 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of P-protein of the glycine cleavage system: implications for nonketotic hyperglycinemia
Embo J., 24, 2005
|
|
5XXT
 
 | | GDP-microtubule complexed with nucleotide-free KIF5C | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Morikawa, M, Shigematsu, H, Nitta, R, Hirokawa, N. | | 登録日 | 2017-07-05 | | 公開日 | 2018-10-10 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (5.35 Å) | | 主引用文献 | Kinesin-binding-triggered conformation switching of microtubules contributes to polarized transport
J. Cell Biol., 217, 2018
|
|
5XXW
 
 | | GDP-microtubule complexed with KIF5C in ATP state | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Morikawa, M, Shigematsu, H, Nitta, R, Hirokawa, N. | | 登録日 | 2017-07-05 | | 公開日 | 2018-10-10 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Kinesin-binding-triggered conformation switching of microtubules contributes to polarized transport
J. Cell Biol., 217, 2018
|
|
5X2L
 
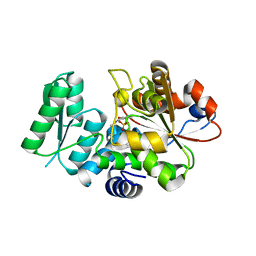 | | Crystal Structure of Human Serine Racemase | | 分子名称: | MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, Serine racemase | | 著者 | Obita, T, Matsumoto, K, Mori, H, Toyooka, N, Mizuguchi, M. | | 登録日 | 2017-02-01 | | 公開日 | 2018-01-31 | | 最終更新日 | 2021-02-24 | | 実験手法 | X-RAY DIFFRACTION (1.806 Å) | | 主引用文献 | Design, synthesis, and evaluation of novel inhibitors for wild-type human serine racemase.
Bioorg. Med. Chem. Lett., 2017
|
|
1CPQ
 
 | | CYTOCHROME C' FROM RHODOPSEUDOMONAS CAPSULATA | | 分子名称: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tahirov, T.H, Misaki, S, Meyer, T.E, Cusanovich, M.A, Higuchi, Y, Yasuoka, N. | | 登録日 | 1995-08-14 | | 公開日 | 1996-12-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | High-resolution crystal structures of two polymorphs of cytochrome c' from the purple phototrophic bacterium rhodobacter capsulatus.
J.Mol.Biol., 259, 1996
|
|
5XJB
 
 | | The Crystal Structure of the Minimal Core Domain of the Microtubule Depolymerizer KIF2C Complexed with ADP-Mg-BeFx | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Kinesin-like protein KIF2C, ... | | 著者 | Ogawa, T, Jiang, X, Hirokawa, N. | | 登録日 | 2017-04-30 | | 公開日 | 2017-09-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Mechanism of Catalytic Microtubule Depolymerization via KIF2-Tubulin Transitional Conformation
Cell Rep, 20, 2017
|
|
2HXF
 
 | |
5XJA
 
 | | The Crystal Structure of the Minimal Core Domain of the Microtubule Depolymerizer KIF2C Complexed with ADP-Mg-AlFx | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Kinesin-like protein KIF2C, ... | | 著者 | Ogawa, T, Jiang, X, Hirokawa, N. | | 登録日 | 2017-04-30 | | 公開日 | 2017-09-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.43 Å) | | 主引用文献 | Mechanism of Catalytic Microtubule Depolymerization via KIF2-Tubulin Transitional Conformation
Cell Rep, 20, 2017
|
|
