3ATZ
 
 | | Crystal structure of TcOYE with pHBA | | Descriptor: | FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZALDEHYDE, Prostaglandin F2a synthase | | Authors: | Okamoto, N, Yamaguchi, K, Mizohata, E, Tokuoka, K, Uchiyama, N, Sugiyama, S, Matsumura, H, Inaka, K, Urade, Y, Inoue, T. | | Deposit date: | 2011-01-26 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insight into the stereoselective production of PGF2(alpha) by Old Yellow Enzyme from Trypanosoma cruzi
J.Biochem., 150, 2011
|
|
1WP4
 
 | |
2IT1
 
 | |
2IT2
 
 | |
2IT3
 
 | |
1WW8
 
 | |
1WL8
 
 | |
1J3W
 
 | |
1WXQ
 
 | |
1WU2
 
 | |
1X1E
 
 | |
1J2W
 
 | | Tetrameric Structure of aldolase from Thermus thermophilus HB8 | | Descriptor: | Aldolase protein | | Authors: | Lokanath, N.K, Shiromizu, I, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2ZKT
 
 | |
1UG6
 
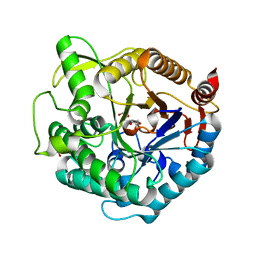 | | Structure of beta-glucosidase at atomic resolution from thermus thermophilus HB8 | | Descriptor: | GLYCEROL, beta-glycosidase | | Authors: | Lokanath, N.K, Shiromizu, I, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-12 | | Release date: | 2003-06-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structure of Beta-Glucosidase at Atomic Resolution from Thermus Thermophilus Hb8
To be Published
|
|
2ZTI
 
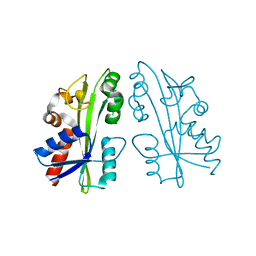 | |
1V7R
 
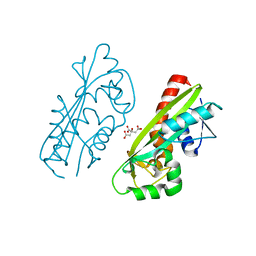 | |
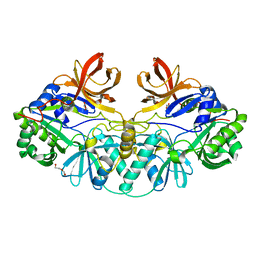
1V5V
 
 | |
1V9S
 
 | |
1V9N
 
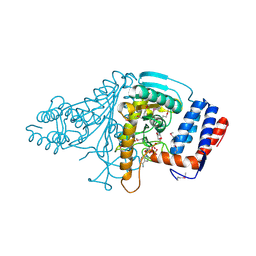 | |
1UB3
 
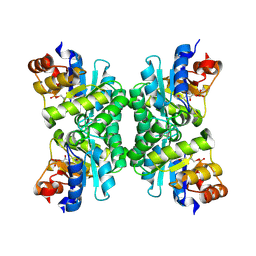 | | Crystal Structure of Tetrameric Structure of Aldolase from thermus thermophilus HB8 | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, Aldolase protein | | Authors: | Lokanath, N.K, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-28 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1VE3
 
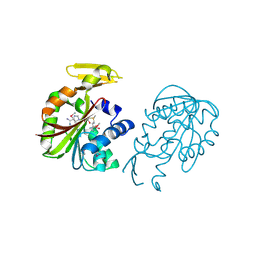 | | Crystal structure of PH0226 protein from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYLMETHIONINE, hypothetical protein PH0226 | | Authors: | Lokanath, N.K, Yamamoto, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-26 | | Release date: | 2005-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase from Pyrococcus horikoshii.
Acta Crystallogr.,Sect.F, 73, 2017
|
|
2CVZ
 
 | |
2DVM
 
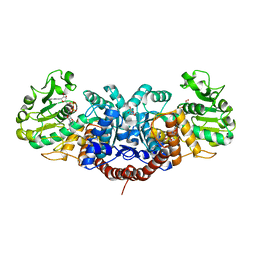 | |
2CUK
 
 | |
2D16
 
 | |
