3GNE
 
 | | Crystal structure of alginate lyase vAL-1 from Chlorella virus | | Descriptor: | CITRATE ANION, GLYCEROL, VAL-1 | | Authors: | Ogura, K, Yamasaki, M, Hashidume, T, Yamada, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-03-17 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action
J.Biol.Chem., 284, 2009
|
|
3IM0
 
 | | Crystal structure of Chlorella virus vAL-1 soaked in 200mM D-glucuronic acid, 10% PEG-3350, and 200mM glycine-NaOH (pH 10.0) | | Descriptor: | VAL-1, beta-D-glucopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Hashidume, T, Yamada, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-08-08 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action
J.Biol.Chem., 284, 2009
|
|
1K1Z
 
 | | Solution structure of N-terminal SH3 domain mutant(P33G) of murine Vav | | Descriptor: | vav | | Authors: | Ogura, K, Nagata, K, Horiuchi, M, Ebisui, E, Hasuda, T, Yuzawa, S, Nishida, M, Hatanaka, H, Inagaki, F. | | Deposit date: | 2001-09-26 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of N-terminal SH3 domain of Vav and the recognition site for Grb2 C-terminal SH3 domain
J.BIOMOL.NMR, 22, 2002
|
|
1WLP
 
 | | Solution Structure Of The P22Phox-P47Phox Complex | | Descriptor: | Cytochrome b-245 light chain, Neutrophil cytosol factor 1 | | Authors: | Ogura, K, Torikai, S, Saikawa, K, Yuzawa, S, Sumimoto, H, Inagaki, F. | | Deposit date: | 2004-06-29 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the tandem Src homology 3 domains of p47phox complexed with a p22phox-derived proline-rich peptide
J.Biol.Chem., 281, 2006
|
|
1QG1
 
 | |
1X0N
 
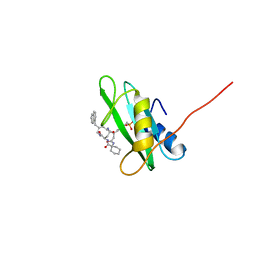 | | NMR structure of growth factor receptor binding protein SH2 domain complexed with the inhibitor | | Descriptor: | 4-[(10S,14S,18S)-18-(2-AMINO-2-OXOETHYL)-14-(1-NAPHTHYLMETHYL)-8,17,20-TRIOXO-7,16,19-TRIAZASPIRO[5.14]ICOS-11-EN-10-YL]BENZYLPHOSPHONIC ACID, Growth factor receptor-bound protein 2 | | Authors: | Ogura, K, Shiga, T, Yuzawa, S, Yokochi, M, Burke, T.R, Inagaki, F. | | Deposit date: | 2005-03-24 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of growth factor receptor binding protein SH2 domain complexed with the inhibitor
To be Published
|
|
2LHI
 
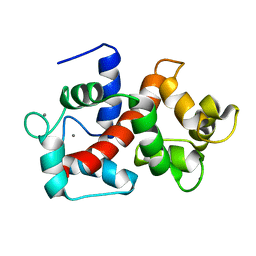 | | Solution structure of Ca2+/CNA1 peptide-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin,Serine/threonine-protein phosphatase 2B catalytic subunit A1 | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
2LHH
 
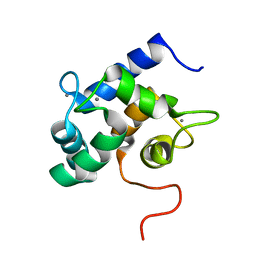 | | Solution structure of Ca2+-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
2Z42
 
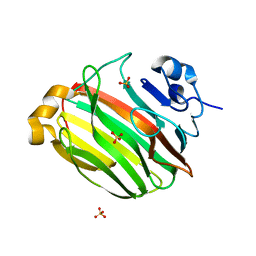 | | Crystal Structure of Family 7 Alginate Lyase A1-II' from Sphingomonas sp. A1 | | Descriptor: | Alginate lyase, SULFATE ION | | Authors: | Ogura, K, Yamasaki, M, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2007-06-12 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate Recognition by Family 7 Alginagte Lyase from Sphingomonas sp. A1
To be published
|
|
2ZAB
 
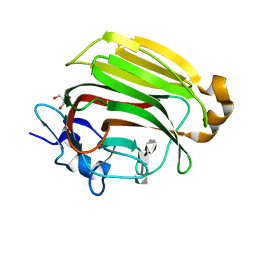 | | Crystal Structure of Family 7 Alginate Lyase A1-II' Y284F in Cmplex with Product (GGG) | | Descriptor: | Alginate lyase, GLYCEROL, alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2ZAC
 
 | | Crystal Structure of Family 7 Alginate Lyase A1-II' Y284F in Complex with Product (MMG) | | Descriptor: | Alginate lyase, GLYCEROL, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
3A0N
 
 | | Crystal structure of D-glucuronic acid-bound alginate lyase vAL-1 from Chlorella virus | | Descriptor: | VAL-1, beta-D-glucopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Hashidume, T, Yamada, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-03-23 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action
J.Biol.Chem., 284, 2009
|
|
2ZAA
 
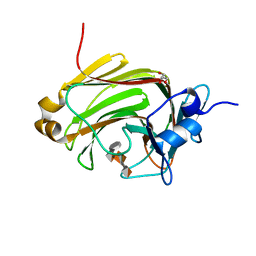 | | Crystal Structure of Family 7 Alginate Lyase A1-II' H191N/Y284F in Complex with Substrate (GGMG) | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, Alginate lyase, GLYCEROL | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2ZA9
 
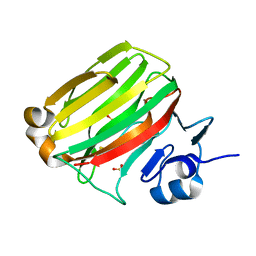 | | Crystal Structure of Alginate lyase A1-II' N141C/N199C | | Descriptor: | Alginate lyase, SULFATE ION | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2YZ0
 
 | |
2KFJ
 
 | | Solution structure of the loop deletion mutant of PB1 domain of Cdc24p | | Descriptor: | Cell division control protein 24 | | Authors: | Ogura, K, Tandai, T, Yoshinaga, S, Kobashigawa, Y, Kumeta, H, Inagaki, F. | | Deposit date: | 2009-02-22 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the heterodimer of Bem1 and Cdc24 PB1 domains from Saccharomyces cerevisiae
J.Biochem., 146, 2009
|
|
8YRI
 
 | | Crystal structure of sugar phosphotransferase system EIIB component CPF_0401 from Clostridium perfringens | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PTS system, mannose/fructose/sorbose family, ... | | Authors: | Hirayama, Y, Oiki, S, Mikami, B, Ogura, K, Hashimoto, W. | | Deposit date: | 2024-03-21 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of sugar phosphotransferase system EIIB component CPF_0401 from Clostridium perfringens
To Be Published
|
|
1GCQ
 
 | | CRYSTAL STRUCTURE OF VAV AND GRB2 SH3 DOMAINS | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
1GCP
 
 | | CRYSTAL STRUCTURE OF VAV SH3 DOMAIN | | Descriptor: | VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
4F13
 
 | | Alginate lyase A1-III Y246F complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-06 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F10
 
 | | Alginate lyase A1-III H192A complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-05 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4E1Y
 
 | | Alginate lyase A1-III H192A apo form | | Descriptor: | Alginate lyase | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-03-07 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2CWS
 
 | | Crystal structure at 1.0 A of alginate lyase A1-II', a member of polysaccharide lyase family-7 | | Descriptor: | GLYCEROL, SULFATE ION, alginate lyase A1-II' | | Authors: | Yamasaki, M, Ogura, K, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2005-06-25 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A Structural Basis for Depolymerization of Alginate by Polysaccharide Lyase Family-7
J.Mol.Biol., 352, 2005
|
|
1AK7
 
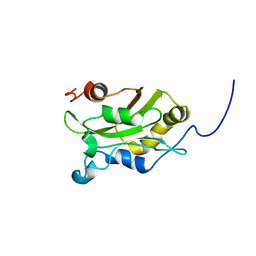 | | DESTRIN, NMR, 20 STRUCTURES | | Descriptor: | DESTRIN | | Authors: | Hatanaka, H, Moriyama, K, Ogura, K, Ichikawa, S, Yahara, I, Inagaki, F. | | Deposit date: | 1997-05-29 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure of destrin and structural similarity between two actin-regulating protein families.
Cell(Cambridge,Mass.), 85, 1996
|
|
1AK6
 
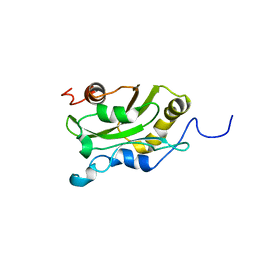 | | DESTRIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DESTRIN | | Authors: | Hatanaka, H, Moriyama, K, Ogura, K, Ichikawa, S, Yahara, I, Inagaki, F. | | Deposit date: | 1997-05-29 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure of destrin and structural similarity between two actin-regulating protein families.
Cell(Cambridge,Mass.), 85, 1996
|
|
