8RSA
 
 | |
6PAE
 
 | |
6NXA
 
 | | ECAII(D90T,K162T) MUTANT AT PH 7 | | Descriptor: | ACETIC ACID, GLYCEROL, L-asparaginase 2 | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2019-02-08 | | Release date: | 2019-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Opportunistic complexes of E. coli L-asparaginases with citrate anions.
Sci Rep, 9, 2019
|
|
8PTI
 
 | | CRYSTAL STRUCTURE OF A Y35G MUTANT OF BOVINE PANCREATIC TRYPSIN INHIBITOR | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR | | Authors: | Housset, D, Kim, K.-S, Fuchs, J, Woodward, C, Wlodawer, A. | | Deposit date: | 1990-12-17 | | Release date: | 1991-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a Y35G mutant of bovine pancreatic trypsin inhibitor.
J.Mol.Biol., 220, 1991
|
|
6NX8
 
 | |
5D9D
 
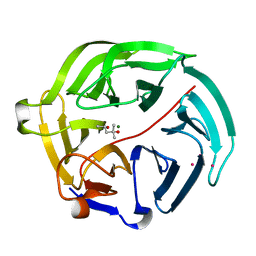 | | Luciferin-regenerating enzyme solved by SAD using synchrotron radiation at room temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
3LJC
 
 | | Crystal structure of Lon N-terminal domain. | | Descriptor: | ATP-dependent protease La | | Authors: | Li, M, Gustchina, A, Dauter, Z, Wlodawer, A. | | Deposit date: | 2010-01-26 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the N-terminal fragment of Escherichia coli Lon protease
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5E0S
 
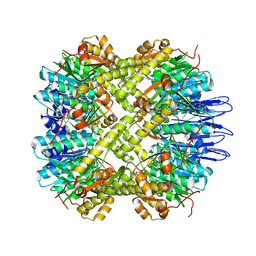 | | crystal structure of the active form of the proteolytic complex clpP1 and clpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2 | | Authors: | LI, M, Wlodawer, A, Maurizi, M. | | Deposit date: | 2015-09-29 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Functional Properties of the Active Form of the Proteolytic Complex, ClpP1P2, from Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
5DZK
 
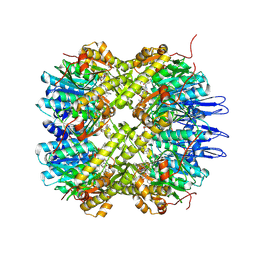 | | Crystal structure of the active form of the proteolytic complex clpP1 and clpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2, BEZ-LEU-LEU | | Authors: | LI, M, Wlodawer, A, Maurizi, M. | | Deposit date: | 2015-09-25 | | Release date: | 2016-02-17 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structure and Functional Properties of the Active Form of the Proteolytic Complex, ClpP1P2, from Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
6X5M
 
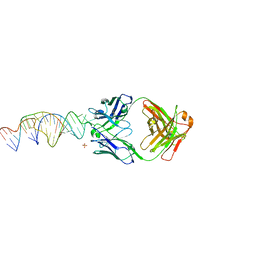 | | Crystal structure of a stabilized PAN ENE bimolecular triplex with a GC-clamped polyA tail, in complex with Fab-BL-3,6. | | Descriptor: | Heavy chain Fab Bl-3 6, Light chain Fab BL-3 6, SULFATE ION, ... | | Authors: | Swain, M, Li, M, Wlodawer, A, Le Grice, S.F.J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamic bulge nucleotides in the KSHV PAN ENE triple helix provide a unique binding platform for small molecule ligands.
Nucleic Acids Res., 49, 2021
|
|
7JOD
 
 | | Crystal structure of BbKI complexed with Human Kallikrein 4 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Kallikrein 4 (Prostase, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-06 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7JOW
 
 | | Crystal structure of BbKI complexed with Human Kallikrein 4 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Kallikrein 4 (Prostase, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-07 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7JOE
 
 | | Crystal structure of BbKI complexed with Human Kallikrein 4 | | Descriptor: | Kallikrein 4 (Prostase, enamel matrix, prostate), ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-06 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI
Acta Crystallogr.,Sect.D, 77, 2021
|
|
9HFQ
 
 | | Cryo-EM structure of human CDADC1 inactive mutant (E400A): trimer without a ligand | | Descriptor: | Cytidine and dCMP deaminase domain-containing protein 1, ZINC ION | | Authors: | Slyvka, A, Rathore, I, Yang, R, Kanai, T, Lountos, G, Wang, Z, Skowronek, K, Czarnocki-Cieciura, M, Wlodawer, A, Bochtler, M. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structure of human CDADC1 inactive mutant (E400A): trimer without a ligand
To Be Published, 2025
|
|
9HFR
 
 | | Cryo-EM structure of human CDADC1 inactive mutant (E400A): hexamer without a ligand | | Descriptor: | Cytidine and dCMP deaminase domain-containing protein 1, ZINC ION | | Authors: | Slyvka, A, Rathore, I, Yang, R, Kanai, T, Lountos, G, Wang, Z, Skowronek, K, Czarnocki-Cieciura, M, Wlodawer, A, Bochtler, M. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of human CDADC1 inactive mutant (E400A): trimer without a ligand
To Be Published, 2025
|
|
9HFT
 
 | | Cryo-EM structure of human CDADC1 inactive mutant (E400A): hexamer with 5mdCTP bound in the active site | | Descriptor: | Cytidine and dCMP deaminase domain-containing protein 1, ZINC ION, [[(2~{R},3~{S},5~{R})-5-(4-azanyl-5-methyl-2-oxidanylidene-pyrimidin-1-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Slyvka, A, Rathore, I, Yang, R, Kanai, T, Lountos, G, Wang, Z, Skowronek, K, Czarnocki-Cieciura, M, Wlodawer, A, Bochtler, M. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of human CDADC1 inactive mutant (E400A): trimer without a ligand
To Be Published, 2025
|
|
5T55
 
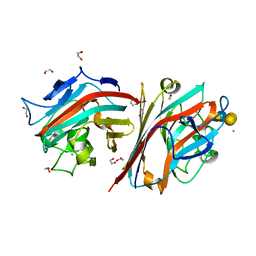 | | LECTIN FROM BAUHINIA FORFICATA IN COMPLEX WITH GLOBOTETRAOSE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2016-08-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural analysis and unique molecular recognition properties of a Bauhinia forficata lectin that inhibits cancer cell growth.
FEBS J., 284, 2017
|
|
5T5L
 
 | | LECTIN FROM BAUHINIA FORFICATA IN COMPLEX WITH TN-PEPTIDE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, ... | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2016-08-31 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural analysis and unique molecular recognition properties of a Bauhinia forficata lectin that inhibits cancer cell growth.
FEBS J., 284, 2017
|
|
5T52
 
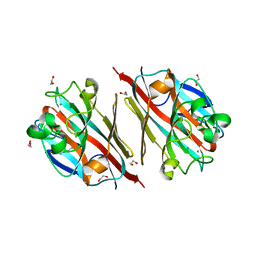 | | LECTIN FROM BAUHINIA FORFICATA IN COMPLEX WITH GALNAC | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2016-08-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis and unique molecular recognition properties of a Bauhinia forficata lectin that inhibits cancer cell growth.
FEBS J., 284, 2017
|
|
5T5J
 
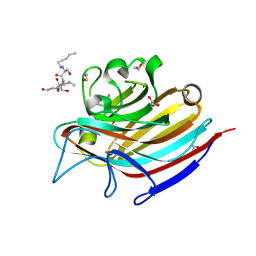 | | LECTIN FROM BAUHINIA FORFICATA IN COMPLEX WITH TN-PEPTIDE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, ... | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2016-08-31 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis and unique molecular recognition properties of a Bauhinia forficata lectin that inhibits cancer cell growth.
FEBS J., 284, 2017
|
|
5T5P
 
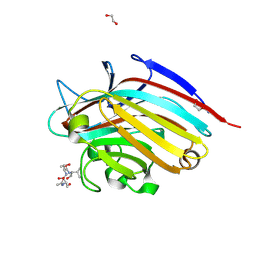 | | LECTIN FROM BAUHINIA FORFICATA IN COMPLEX WITH TN-PEPTIDE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, ... | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2016-08-31 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural analysis and unique molecular recognition properties of a Bauhinia forficata lectin that inhibits cancer cell growth.
FEBS J., 284, 2017
|
|
6RIZ
 
 | | Crystal structure of gp41-1 intein (C1A, F65W, D107C) | | Descriptor: | gp41-1 intein | | Authors: | Beyer, H.M, Lountos, G.T, Mikula, M.K, Wlodawer, A, Iwai, H. | | Deposit date: | 2019-04-25 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Convergence of the Hedgehog/Intein Fold in Different Protein Splicing Mechanisms.
Int J Mol Sci, 21, 2020
|
|
9HFS
 
 | | Cryo-EM structure of human CDADC1 inactive mutant (E400A): hexamer with dCTP bound in the active site | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Cytidine and dCMP deaminase domain-containing protein 1, ZINC ION | | Authors: | Slyvka, A, Rathore, I, Yang, R, Kanai, T, Lountos, G, Wang, Z, Skowronek, K, Czarnocki-Cieciura, M, Wlodawer, A, Bochtler, M. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of human CDADC1 inactive mutant (E400A): trimer without a ligand
To Be Published, 2025
|
|
5T5O
 
 | | LECTIN FROM BAUHINIA FORFICATA IN COMPLEX WITH TN-PEPTIDE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, Lectin, ... | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2016-08-31 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis and unique molecular recognition properties of a Bauhinia forficata lectin that inhibits cancer cell growth.
FEBS J., 284, 2017
|
|
3NR6
 
 | | Crystal structure of xenotropic murine leukemia virus-related virus (XMRV) protease | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Protease p14 | | Authors: | Lubkowski, J, Li, M, Gustchina, A, Zhou, D, Dauter, Z, Wlodawer, A. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of XMRV protease differs from the structures of other retropepsins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
