8JLD
 
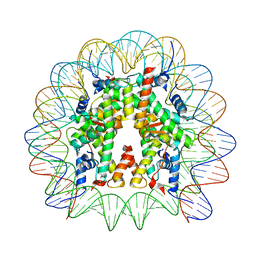 | | Cryo-EM structure of the 145 bp human nucleosome containing acetylated H3 tail | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
8JLA
 
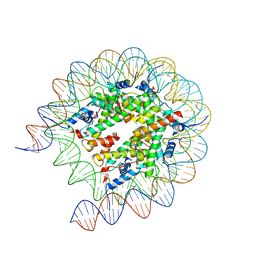 | | Cryo-EM structure of the human nucleosome lacking N-terminal region of H2A, H2B, H3, and H4 | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
6A3K
 
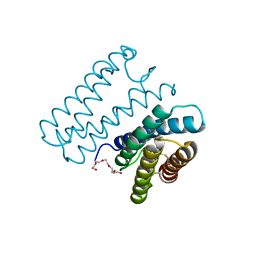 | | Crystal structure of cytochrome c' from Shewanella benthica DB6705 | | Descriptor: | Cytochrome c, HEME C, PENTAETHYLENE GLYCOL | | Authors: | Suka, A, Oki, H, Kato, Y, Kawahara, K, Ohkubo, T, Maruno, T, Kobayashi, Y, Fujii, S, Wakai, S, Sambongi, Y. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-12 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Stability of cytochromes c' from psychrophilic and piezophilic Shewanella species: implications for complex multiple adaptation to low temperature and high hydrostatic pressure.
Extremophiles, 23, 2019
|
|
6LUG
 
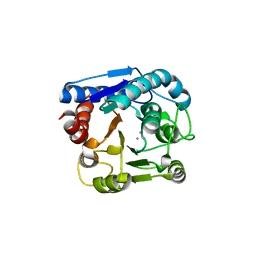 | | Crystal structure of N(omega)-hydroxy-L-arginine hydrolase | | Descriptor: | MANGANESE (II) ION, N(omega)-hydroxy-L-arginine amidinohydrolase | | Authors: | Oda, K, Matoba, Y. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an Nomega-hydroxy-L-arginine hydrolase found in the D-cycloserine biosynthetic pathway.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5B6Q
 
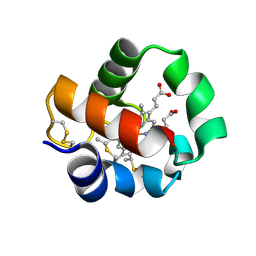 | | Crystal structure of monomeric cytochrome c5 from Shewanella violacea | | Descriptor: | HEME C, IMIDAZOLE, Soluble cytochrome cA | | Authors: | Masanari, M, Fujii, S, Kawahara, K, Oki, H, Tsujino, H, Maruno, T, Kobayashi, Y, Ohkubo, T, Nishiyama, M, Harada, Y, Wakai, S, Sambongi, Y. | | Deposit date: | 2016-06-01 | | Release date: | 2016-10-19 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Comparative study on stabilization mechanism of monomeric cytochrome c5 from deep-sea piezophilic Shewanella violacea
Biosci.Biotechnol.Biochem., 2016
|
|
5B3I
 
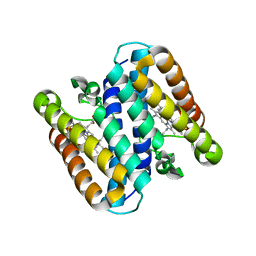 | | Homo-dimeric structure of cytochrome c' from Thermophilic Hydrogenophilus thermoluteolus | | Descriptor: | Cytochrome c prime, HEME C | | Authors: | Fujii, S, Oki, H, Kawahara, K, Yamane, D, Yamanaka, M, Maruno, T, Kobayashi, Y, Masanari, M, Wakai, S, Nishihara, H, Ohkubo, T, Sambongi, Y. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and functional insights into thermally stable cytochrome c' from a thermophile
Protein Sci., 26, 2017
|
|
5AX6
 
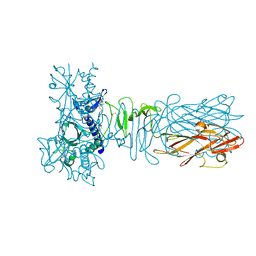 | | The crystal structure of CofB, the minor pilin subunit of CFA/III from human enterotoxigenic Escherichia coli. | | Descriptor: | ACETATE ION, CofB | | Authors: | Kawahara, K, Oki, K, Fukaksua, F, Maruno, T, Kobayashi, Y, Daisuke, M, Taniguchi, T, Honda, T, Iida, T, Nakamura, S, Ohkubo, T. | | Deposit date: | 2015-07-16 | | Release date: | 2016-03-09 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Homo-trimeric Structure of the Type IVb Minor Pilin CofB Suggests Mechanism of CFA/III Pilus Assembly in Human Enterotoxigenic Escherichia coli
J.Mol.Biol., 428, 2016
|
|
8IUS
 
 | |
8IUU
 
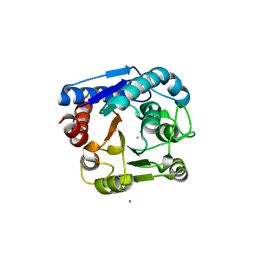 | |
8IUV
 
 | |
8IUW
 
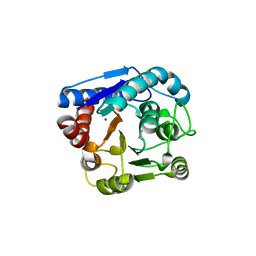 | |
7DC0
 
 | |
7DC4
 
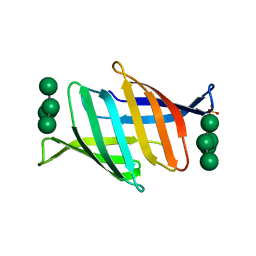 | | Crystal structure of glycan-bound Pseudomonas taiwanensis lectin | | Descriptor: | Lectin, SULFATE ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose | | Authors: | Oda, K, Matoba, Y. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Lectins engineered to favor a glycan-binding conformation have enhanced antiviral activity.
J.Biol.Chem., 296, 2021
|
|
7CIT
 
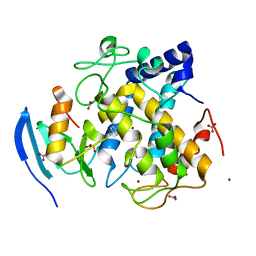 | |
7CIY
 
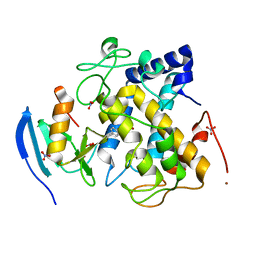 | |
7VDY
 
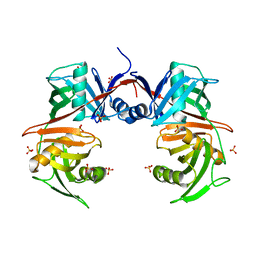 | | Crystal structure of O-ureidoserine racemase | | Descriptor: | O-ureido-serine racemase, SULFATE ION | | Authors: | Oda, K, Matoba, Y. | | Deposit date: | 2021-09-07 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of O-ureidoserine racemase found in the d-cycloserine biosynthetic pathway.
Proteins, 90, 2022
|
|
7EUN
 
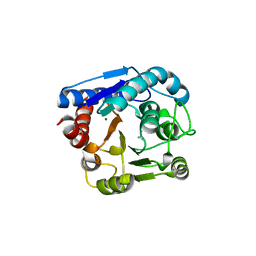 | |
7EUQ
 
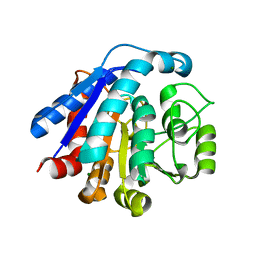 | |
7EUL
 
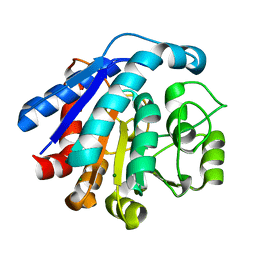 | |
7EUK
 
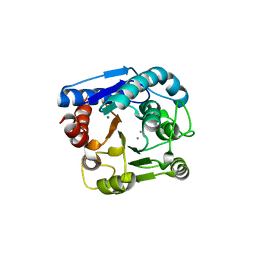 | |
3VX7
 
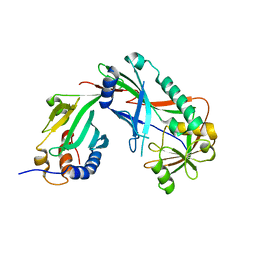 | | Crystal structure of Kluyveromyces marxianus Atg7NTD-Atg10 complex | | Descriptor: | E1, E2 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VX6
 
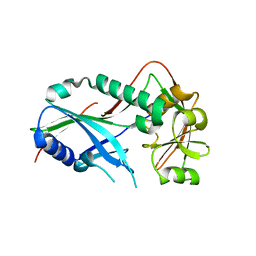 | | Crystal structure of Kluyveromyces marxianus Atg7NTD | | Descriptor: | E1 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3WJ4
 
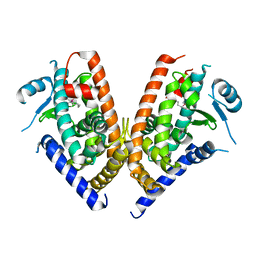 | | Crystal structure of PPARgamma ligand binding domain in complex with tributyltin | | Descriptor: | Peroxisome proliferator-activated receptor gamma, tributylstannanyl | | Authors: | Harada, S, Hiromori, Y, Fukakusa, S, Kawahara, K, Nakamura, S, Noda, M, Uchiyama, S, Fukui, K, Nishikawa, J, Nagase, H, Kobayashi, Y, Ohkubo, T, Yoshida, T, Nakanishi, T. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for PPARgamma transactivation by endocrine disrupting organotin compounds
To be Published
|
|
3WJ5
 
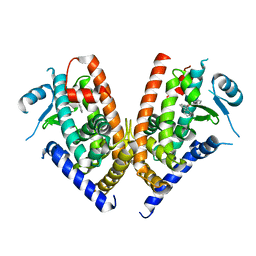 | | Crystal structure of PPARgamma ligand binding domain in complex with triphenyltin | | Descriptor: | Peroxisome proliferator-activated receptor gamma, triphenylstannanyl | | Authors: | Harada, S, Hiromori, Y, Fukakusa, S, Kawahara, K, Nakamura, S, Noda, M, Uchiyama, S, Fukui, K, Nishikawa, J, Nagase, H, Kobayashi, Y, Ohkubo, T, Yoshida, T, Nakanishi, T. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for PPARgamma transactivation by endocrine disrupting organotin compounds
To be Published
|
|
3VOR
 
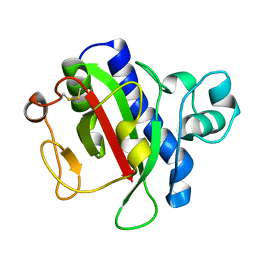 | | Crystal Structure Analysis of the CofA | | Descriptor: | CFA/III pilin | | Authors: | Fukakusa, S, Kawahara, K, Nakamura, S, Iwasita, T, Baba, S, Nishimura, M, Kobayashi, Y, Honda, T, Iida, T, Taniguchi, T, Ohkubo, T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-09-26 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structure of the CFA/III major pilin subunit CofA from human enterotoxigenic Escherichia coli determined at 0.90 A resolution by sulfur-SAD phasing
Acta Crystallogr.,Sect.D, 68, 2012
|
|
