2KE4
 
 | |
2KBT
 
 | | Attachment of an NMR-invisible solubility enhancement tag (INSET) using a sortase-mediated protein ligation method | | 分子名称: | Proto-oncogene vav,Immunoglobulin G-binding protein G | | 著者 | Kumeta, H, Kobashigawa, Y, Ogura, K, Inagaki, F. | | 登録日 | 2008-12-07 | | 公開日 | 2009-02-03 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Attachment of an NMR-invisible solubility enhancement tag using a sortase-mediated protein ligation method
J.Biomol.Nmr, 43, 2009
|
|
1WPK
 
 | | Methylated Form of N-terminal Transcriptional Regulator Domain of Escherichia Coli Ada Protein | | 分子名称: | ADA regulatory protein, ZINC ION | | 著者 | Takinowaki, H, Matsuda, Y, Yoshida, T, Kobayashi, Y, Ohkubo, T. | | 登録日 | 2004-09-07 | | 公開日 | 2005-09-13 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the methylated form of the N-terminal 16-kDa domain of Escherichia coli Ada protein
Protein Sci., 15, 2006
|
|
1Y1C
 
 | | Solution structure of Anemonia elastase inhibitor analogue | | 分子名称: | Elastase inhibitor | | 著者 | Hemmi, H, Kumazaki, T, Yoshizawa-Kumagaye, K, Nishiuchi, Y, Yoshida, T, Ohkubo, T, Kobayashi, Y. | | 登録日 | 2004-11-18 | | 公開日 | 2005-07-19 | | 最終更新日 | 2021-11-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and Functional Study of an Anemonia Elastase Inhibitor, a "Nonclassical" Kazal-Type Inhibitor from Anemonia sulcata
Biochemistry, 44, 2005
|
|
3WWT
 
 | |
1Y1B
 
 | | Solution structure of Anemonia elastase inhibitor | | 分子名称: | Elastase inhibitor | | 著者 | Hemmi, H, Kumazaki, T, Yoshizawa-Kumagaye, K, Nishiuchi, Y, Yoshida, T, Ohkubo, T, Kobayashi, Y. | | 登録日 | 2004-11-18 | | 公開日 | 2005-07-19 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and Functional Study of an Anemonia Elastase Inhibitor, a "Nonclassical" Kazal-Type Inhibitor from Anemonia sulcata
Biochemistry, 44, 2005
|
|
3VVL
 
 | | Crystal structure of L-serine-O-acetyltransferase found in D-cycloserine biosynthetic pathway | | 分子名称: | Homoserine O-acetyltransferase | | 著者 | Oda, K, Matoba, Y, Kumagai, T, Noda, M, Sugiyama, M. | | 登録日 | 2012-07-26 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Crystallographic study to determine the substrate specificity of an L-serine-acetylating enzyme found in the D-cycloserine biosynthetic pathway
J.Bacteriol., 195, 2013
|
|
3VVM
 
 | | Crystal structure of G52A-P55G mutant of L-serine-O-acetyltransferase found in D-cycloserine biosynthetic pathway | | 分子名称: | Homoserine O-acetyltransferase | | 著者 | Oda, K, Matoba, Y, Kumagai, T, Noda, M, Sugiyama, M. | | 登録日 | 2012-07-26 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystallographic study to determine the substrate specificity of an L-serine-acetylating enzyme found in the D-cycloserine biosynthetic pathway
J.Bacteriol., 195, 2013
|
|
3FHP
 
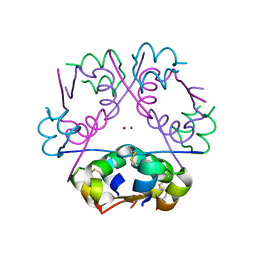 | | A neutron crystallographic analysis of a porcine 2Zn insulin at 2.0 A resolution | | 分子名称: | Insulin, ZINC ION | | 著者 | Iwai, W, Kurihara, K, Yamada, T, Kobayashi, Y, Ohnishi, Y, Tanaka, I, Takahashi, H, Niimura, N. | | 登録日 | 2008-12-09 | | 公開日 | 2009-10-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | NEUTRON DIFFRACTION (2 Å) | | 主引用文献 | A neutron crystallographic analysis of T6 porcine insulin at 2.1 A resolution
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3B2C
 
 | | Crystal structure of the collagen triple helix model [{PRO-HYP(R)-GLY}4-{HYP(S)-Pro-GLY}2-{PRO-HYP(R)-GLY}4]3 | | 分子名称: | Collagen-like peptide | | 著者 | Motooka, D, Kawahara, K, Nakamura, S, Doi, M, Nishi, Y, Nishiuchi, Y, Nakazawa, T, Yoshida, T, Ohkubo, T, Kobayashi, Y, Kang, Y.K, Uchiyama, S. | | 登録日 | 2011-07-26 | | 公開日 | 2012-04-04 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | The triple helical structure and stability of collagen model peptide with 4(S)-hydroxyprolyl-pro-gly units
Biopolymers, 98, 2011
|
|
1V6R
 
 | | Solution Structure of Endothelin-1 with its C-terminal Folding | | 分子名称: | Endothelin-1 | | 著者 | Takashima, H, Mimura, N, Ohkubo, T, Yoshida, T, Tamaoki, H, Kobayashi, Y. | | 登録日 | 2003-12-03 | | 公開日 | 2004-03-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Distributed Computing and NMR Constraint-Based High-Resolution Structure
Determination: Applied for Bioactive Peptide Endothelin-1 To Determine C-Terminal
Folding
J.Am.Chem.Soc., 126, 2004
|
|
1WKI
 
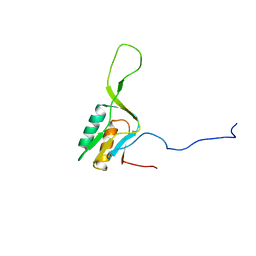 | | solution structure of ribosomal protein L16 from thermus thermophilus HB8 | | 分子名称: | LSU ribosomal protein L16P | | 著者 | Nishimura, M, Yoshida, T, Shirouzu, M, Terada, T, Kuramitsu, S, Yokoyama, S, Ohkubo, T, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-05-31 | | 公開日 | 2004-12-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Ribosomal Protein L16 from Thermus thermophilus HB8
J.Mol.Biol., 344, 2004
|
|
1Y69
 
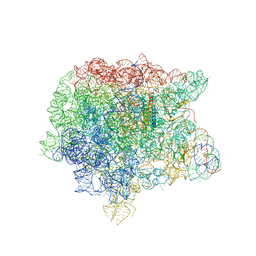 | | RRF domain I in complex with the 50S ribosomal subunit from Deinococcus radiodurans | | 分子名称: | 23S ribosomal RNA, 50S ribosomal protein L16, 50S ribosomal protein L27, ... | | 著者 | Wilson, D.N, Schluenzen, F, Harms, J.M, Yoshida, T, Ohkubo, T, Albrecht, R, Buerger, J, Kobayashi, Y, Fucini, P. | | 登録日 | 2004-12-04 | | 公開日 | 2005-03-01 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.33 Å) | | 主引用文献 | X-ray crystallography on ribosome recycling: mechanism of binding and action of RRF on the 50S ribosomal subunit
EMBO J., 24, 2005
|
|
2RPV
 
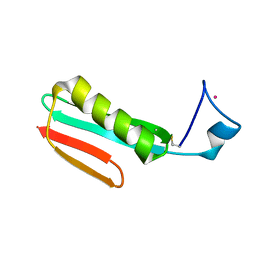 | | Solution Structure of GB1 with LBT probe | | 分子名称: | Immunoglobulin G-binding protein G, LANTHANUM (III) ION | | 著者 | Saio, T, Ogura, K, Yokochi, M, Kobashigawa, Y, Inagaki, F. | | 登録日 | 2008-10-28 | | 公開日 | 2009-09-15 | | 最終更新日 | 2021-11-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Two-point anchoring of a lanthanide-binding peptide to a target protein enhances the paramagnetic anisotropic effect
J.Biomol.Nmr, 44, 2009
|
|
2ZXY
 
 | | Crystal Structure of Cytochrome c555 from Aquifex aeolicus | | 分子名称: | Cytochrome c552, HEME C | | 著者 | Obuchi, M, Kawahara, K, Motooka, D, Nakamura, S, Yamanaka, M, Takeda, T, Uchiyama, S, Kobayashi, Y, Ohkubo, T, Sambongi, Y. | | 登録日 | 2009-01-09 | | 公開日 | 2009-08-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Hyperstability and crystal structure of cytochrome c(555) from hyperthermophilic Aquifex aeolicus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1VFH
 
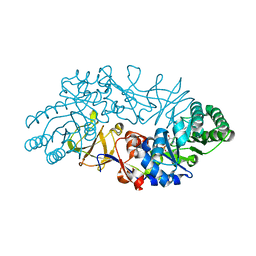 | | Crystal structure of alanine racemase from D-cycloserine producing Streptomyces lavendulae | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, alanine racemase | | 著者 | Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | 登録日 | 2004-04-13 | | 公開日 | 2004-09-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
J.Biol.Chem., 279, 2004
|
|
1BQT
 
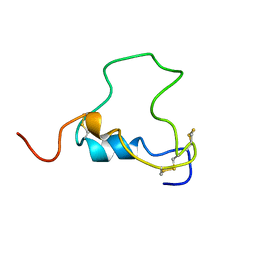 | | THREE-DIMENSIONAL STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR-I (IGF-I) DETERMINED BY 1H-NMR AND DISTANCE GEOMETRY, 6 STRUCTURES | | 分子名称: | INSULIN-LIKE GROWTH FACTOR-I | | 著者 | Sato, A, Nishimura, S, Ohkubo, T, Kyogoku, Y, Koyama, S, Kobayashi, M, Yasuda, T, Kobayashi, Y. | | 登録日 | 1998-08-18 | | 公開日 | 1999-05-18 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of human insulin-like growth factor-I (IGF-I) determined by 1H-NMR and distance geometry.
Int.J.Pept.Protein Res., 41, 1993
|
|
2Z2T
 
 | |
2Z6W
 
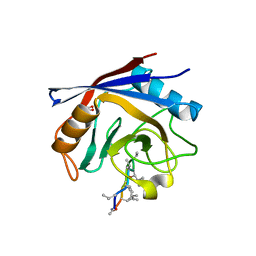 | | Crystal structure of human cyclophilin D in complex with cyclosporin A | | 分子名称: | CITRIC ACID, CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | 著者 | Kajitani, K, Fujihashi, M, Kobayashi, Y, Shimizu, S, Tsujimoto, Y, Miki, K. | | 登録日 | 2007-08-09 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.96 Å) | | 主引用文献 | Crystal Structure of Human Cyclophilin D in Complex with its Inhibitor, Cyclosporin a at 0.96-A Resolution.
Proteins, 70, 2008
|
|
3AHA
 
 | | Crystal structure of the complex between gp41 fragments N36 and C34 mutant N126K/E137Q | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Transmembrane protein gp41 | | 著者 | Izumi, K, Nakamura, S, Nakano, H, Shimura, K, Sakagami, Y, Oishi, S, Uchiyama, S, Ohkubo, T, Kobayashi, Y, Fujii, N, Matsuoka, M, Kodama, E.N. | | 登録日 | 2010-04-22 | | 公開日 | 2010-05-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Characterization of HIV-1 resistance to a fusion inhibitor, N36, derived from the gp41 amino terminal heptad repeat.
Antiviral Res., 2010
|
|
1QTO
 
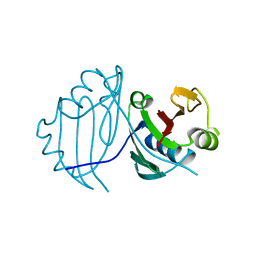 | | 1.5 A CRYSTAL STRUCTURE OF A BLEOMYCIN RESISTANCE DETERMINANT FROM BLEOMYCIN-PRODUCING STREPTOMYCES VERTICILLUS | | 分子名称: | BLEOMYCIN-BINDING PROTEIN | | 著者 | Kawano, Y, Kumagai, T, Muta, K, Matoba, Y, Davies, J, Sugiyama, M. | | 登録日 | 1999-06-28 | | 公開日 | 2000-06-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The 1.5 A crystal structure of a bleomycin resistance determinant from bleomycin-producing Streptomyces verticillus.
J.Mol.Biol., 295, 2000
|
|
1VFT
 
 | | Crystal structure of L-cycloserine-bound form of alanine racemase from D-cycloserine-producing Streptomyces lavendulae | | 分子名称: | CHLORIDE ION, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, alanine racemase | | 著者 | Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | 登録日 | 2004-04-19 | | 公開日 | 2004-09-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
J.Biol.Chem., 279, 2004
|
|
1VFS
 
 | | Crystal structure of D-cycloserine-bound form of alanine racemase from D-cycloserine-producing Streptomyces lavendulae | | 分子名称: | CHLORIDE ION, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, alanine racemase | | 著者 | Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | 登録日 | 2004-04-19 | | 公開日 | 2004-09-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
J.Biol.Chem., 279, 2004
|
|
3AQB
 
 | | M. luteus B-P 26 heterodimeric hexaprenyl diphosphate synthase in complex with magnesium | | 分子名称: | CHLORIDE ION, Component A of hexaprenyl diphosphate synthase, Component B of hexaprenyl diphosphate synthase, ... | | 著者 | Sasaki, D, Fujihashi, M, Okuyama, N, Kobayashi, Y, Noike, M, Koyama, T, Miki, K. | | 登録日 | 2010-10-28 | | 公開日 | 2010-11-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of heterodimeric hexaprenyl diphosphate synthase from Micrococcus luteus B-P 26 reveals that the small subunit is directly involved in the product chain length regulation.
J.Biol.Chem., 286, 2011
|
|
1T7H
 
 | | X-ray structure of [Lys(-2)-Arg(-1)-des(17-21)]-endothelin-1 peptide | | 分子名称: | Endothelin-1 | | 著者 | Hoh, F, Cerdan, R, Kaas, Q, Nishi, Y, Chiche, L, Kubo, S, Chino, N, Kobayashi, Y, Dumas, C, Aumelas, A. | | 登録日 | 2004-05-10 | | 公開日 | 2004-12-21 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | High-resolution X-ray structure of the unexpectedly stable dimer of the [Lys(-2)-Arg(-1)-des(17-21)]endothelin-1 peptide
Biochemistry, 43, 2004
|
|
