6KP3
 
 | |
2KFJ
 
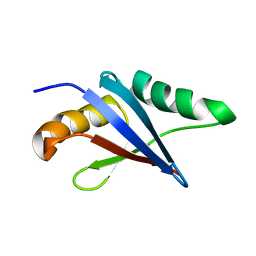 | | Solution structure of the loop deletion mutant of PB1 domain of Cdc24p | | 分子名称: | Cell division control protein 24 | | 著者 | Ogura, K, Tandai, T, Yoshinaga, S, Kobashigawa, Y, Kumeta, H, Inagaki, F. | | 登録日 | 2009-02-22 | | 公開日 | 2009-10-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the heterodimer of Bem1 and Cdc24 PB1 domains from Saccharomyces cerevisiae
J.Biochem., 146, 2009
|
|
2LDR
 
 | |
2LHI
 
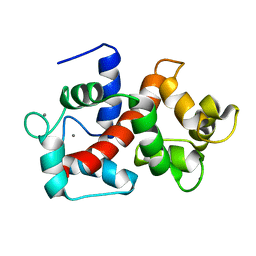 | | Solution structure of Ca2+/CNA1 peptide-bound yCaM | | 分子名称: | CALCIUM ION, Calmodulin,Serine/threonine-protein phosphatase 2B catalytic subunit A1 | | 著者 | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | 登録日 | 2011-08-10 | | 公開日 | 2012-08-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
3FHP
 
 | | A neutron crystallographic analysis of a porcine 2Zn insulin at 2.0 A resolution | | 分子名称: | Insulin, ZINC ION | | 著者 | Iwai, W, Kurihara, K, Yamada, T, Kobayashi, Y, Ohnishi, Y, Tanaka, I, Takahashi, H, Niimura, N. | | 登録日 | 2008-12-09 | | 公開日 | 2009-10-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | NEUTRON DIFFRACTION (2 Å) | | 主引用文献 | A neutron crystallographic analysis of T6 porcine insulin at 2.1 A resolution
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3B2C
 
 | | Crystal structure of the collagen triple helix model [{PRO-HYP(R)-GLY}4-{HYP(S)-Pro-GLY}2-{PRO-HYP(R)-GLY}4]3 | | 分子名称: | Collagen-like peptide | | 著者 | Motooka, D, Kawahara, K, Nakamura, S, Doi, M, Nishi, Y, Nishiuchi, Y, Nakazawa, T, Yoshida, T, Ohkubo, T, Kobayashi, Y, Kang, Y.K, Uchiyama, S. | | 登録日 | 2011-07-26 | | 公開日 | 2012-04-04 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | The triple helical structure and stability of collagen model peptide with 4(S)-hydroxyprolyl-pro-gly units
Biopolymers, 98, 2011
|
|
1VA2
 
 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 2) | | 分子名称: | Transcription factor Sp1, ZINC ION | | 著者 | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | 登録日 | 2004-02-07 | | 公開日 | 2005-02-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1VA3
 
 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 3) | | 分子名称: | Transcription factor Sp1, ZINC ION | | 著者 | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | 登録日 | 2004-02-07 | | 公開日 | 2005-02-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1VA1
 
 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 1) | | 分子名称: | Transcription factor Sp1, ZINC ION | | 著者 | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | 登録日 | 2004-02-07 | | 公開日 | 2005-02-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1T7H
 
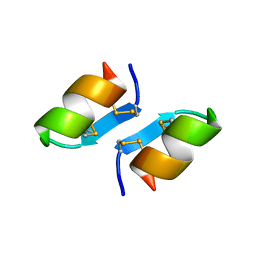 | | X-ray structure of [Lys(-2)-Arg(-1)-des(17-21)]-endothelin-1 peptide | | 分子名称: | Endothelin-1 | | 著者 | Hoh, F, Cerdan, R, Kaas, Q, Nishi, Y, Chiche, L, Kubo, S, Chino, N, Kobayashi, Y, Dumas, C, Aumelas, A. | | 登録日 | 2004-05-10 | | 公開日 | 2004-12-21 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | High-resolution X-ray structure of the unexpectedly stable dimer of the [Lys(-2)-Arg(-1)-des(17-21)]endothelin-1 peptide
Biochemistry, 43, 2004
|
|
3VQI
 
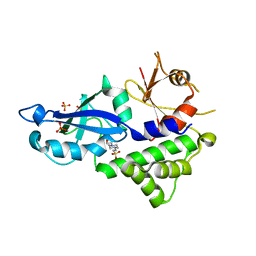 | | Crystal structure of Kluyveromyces marxianus Atg5 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Atg5, SULFATE ION | | 著者 | Yamaguchi, M, Noda, N.N, Yamamoto, H, Shima, T, Kumeta, H, Kobashigawa, Y, Akada, R, Ohsumi, Y, Inagaki, F. | | 登録日 | 2012-03-24 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into atg10-mediated formation of the autophagy-essential atg12-atg5 conjugate
Structure, 20, 2012
|
|
1QTO
 
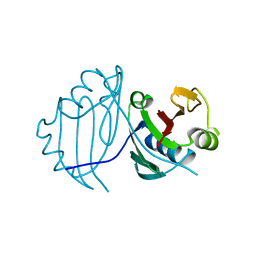 | | 1.5 A CRYSTAL STRUCTURE OF A BLEOMYCIN RESISTANCE DETERMINANT FROM BLEOMYCIN-PRODUCING STREPTOMYCES VERTICILLUS | | 分子名称: | BLEOMYCIN-BINDING PROTEIN | | 著者 | Kawano, Y, Kumagai, T, Muta, K, Matoba, Y, Davies, J, Sugiyama, M. | | 登録日 | 1999-06-28 | | 公開日 | 2000-06-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The 1.5 A crystal structure of a bleomycin resistance determinant from bleomycin-producing Streptomyces verticillus.
J.Mol.Biol., 295, 2000
|
|
6A3L
 
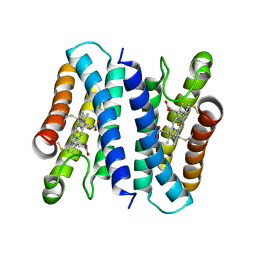 | | Crystal structure of cytochrome c' from Shewanella violacea DSS12 | | 分子名称: | Cytochrome c, HEME C | | 著者 | Suka, A, Oki, H, Kato, Y, Kawahara, K, Ohkubo, T, Maruno, T, Kobayashi, Y, Fujii, S, Wakai, S, Sambongi, Y. | | 登録日 | 2018-06-15 | | 公開日 | 2019-06-12 | | 最終更新日 | 2019-10-02 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Stability of cytochromes c' from psychrophilic and piezophilic Shewanella species: implications for complex multiple adaptation to low temperature and high hydrostatic pressure.
Extremophiles, 23, 2019
|
|
2KE4
 
 | |
2KBT
 
 | | Attachment of an NMR-invisible solubility enhancement tag (INSET) using a sortase-mediated protein ligation method | | 分子名称: | Proto-oncogene vav,Immunoglobulin G-binding protein G | | 著者 | Kumeta, H, Kobashigawa, Y, Ogura, K, Inagaki, F. | | 登録日 | 2008-12-07 | | 公開日 | 2009-02-03 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Attachment of an NMR-invisible solubility enhancement tag using a sortase-mediated protein ligation method
J.Biomol.Nmr, 43, 2009
|
|
1IS1
 
 | | Crystal structure of ribosome recycling factor from Vibrio parahaemolyticus | | 分子名称: | RIBOSOME RECYCLING FACTOR | | 著者 | Nakano, H, Yamaichi, Y, Uchiyama, S, Yoshida, T, Nishina, K, Kato, H, Ohkubo, T, Honda, T, Yamagata, Y, Kobayashi, Y. | | 登録日 | 2001-11-05 | | 公開日 | 2003-06-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure and binding mode of a ribosome recycling factor (RRF) from mesophilic bacterium
J.BIOL.CHEM., 278, 2003
|
|
2Z6W
 
 | | Crystal structure of human cyclophilin D in complex with cyclosporin A | | 分子名称: | CITRIC ACID, CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | 著者 | Kajitani, K, Fujihashi, M, Kobayashi, Y, Shimizu, S, Tsujimoto, Y, Miki, K. | | 登録日 | 2007-08-09 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.96 Å) | | 主引用文献 | Crystal Structure of Human Cyclophilin D in Complex with its Inhibitor, Cyclosporin a at 0.96-A Resolution.
Proteins, 70, 2008
|
|
2RPV
 
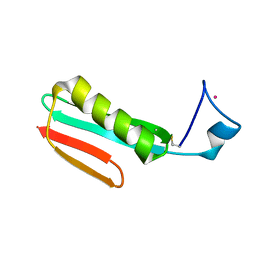 | | Solution Structure of GB1 with LBT probe | | 分子名称: | Immunoglobulin G-binding protein G, LANTHANUM (III) ION | | 著者 | Saio, T, Ogura, K, Yokochi, M, Kobashigawa, Y, Inagaki, F. | | 登録日 | 2008-10-28 | | 公開日 | 2009-09-15 | | 最終更新日 | 2021-11-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Two-point anchoring of a lanthanide-binding peptide to a target protein enhances the paramagnetic anisotropic effect
J.Biomol.Nmr, 44, 2009
|
|
5YQ0
 
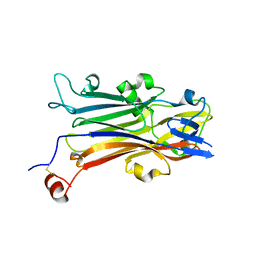 | | Crystal structure of secreted protein CofJ from ETEC. | | 分子名称: | CALCIUM ION, CofJ | | 著者 | Oki, H, Kawahara, K, Maruno, T, Imai, T, Muroga, Y, Fukakusa, S, Iwashita, T, Kobayashi, Y, Matsuda, S, Kodama, T, Iida, T, Yoshida, T, Ohkubo, T, Nakamura, S. | | 登録日 | 2017-11-04 | | 公開日 | 2018-06-27 | | 最終更新日 | 2018-07-25 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Interplay of a secreted protein with type IVb pilus for efficient enterotoxigenicEscherichia colicolonization
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2ZXY
 
 | | Crystal Structure of Cytochrome c555 from Aquifex aeolicus | | 分子名称: | Cytochrome c552, HEME C | | 著者 | Obuchi, M, Kawahara, K, Motooka, D, Nakamura, S, Yamanaka, M, Takeda, T, Uchiyama, S, Kobayashi, Y, Ohkubo, T, Sambongi, Y. | | 登録日 | 2009-01-09 | | 公開日 | 2009-08-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Hyperstability and crystal structure of cytochrome c(555) from hyperthermophilic Aquifex aeolicus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5YPZ
 
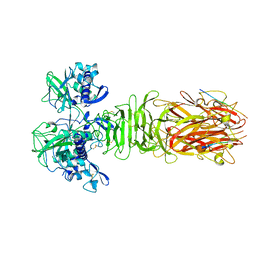 | | Crystal structure of minor pilin CofB from CFA/III complexed with N-terminal peptide fragment of CofJ | | 分子名称: | CofB, CofJ | | 著者 | Oki, H, Kawahara, K, Maruno, T, Imai, T, Muroga, Y, Fukakusa, S, Iwashita, T, Kobayashi, Y, Matsuda, S, Kodama, T, Iida, T, Yoshida, T, Ohkubo, T, Nakamura, S. | | 登録日 | 2017-11-04 | | 公開日 | 2018-06-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.521 Å) | | 主引用文献 | Interplay of a secreted protein with type IVb pilus for efficient enterotoxigenicEscherichia colicolonization.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1JIE
 
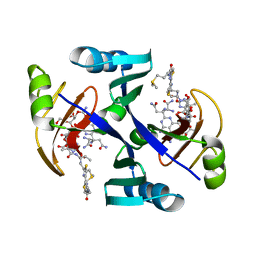 | | Crystal structure of bleomycin-binding protein from bleomycin-producing Streptomyces verticillus complexed with metal-free bleomycin | | 分子名称: | BLEOMYCIN A2, bleomycin-binding protein | | 著者 | Sugiyama, M, Kumagai, T, Hayashida, M, Maruyama, M, Matoba, Y. | | 登録日 | 2001-07-02 | | 公開日 | 2002-02-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The 1.6-A crystal structure of the copper(II)-bound bleomycin complexed with the bleomycin-binding protein from bleomycin-producing Streptomyces verticillus.
J.Biol.Chem., 277, 2002
|
|
1JIF
 
 | | Crystal structure of bleomycin-binding protein from bleomycin-producing Streptomyces verticillus complexed with copper(II)-bleomycin | | 分子名称: | BLEOMYCIN A2, CHLORIDE ION, COPPER (II) ION, ... | | 著者 | Sugiyama, M, Kumagai, T, Hayashida, M, Maruyama, M, Matoba, Y. | | 登録日 | 2001-07-02 | | 公開日 | 2002-02-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The 1.6-A crystal structure of the copper(II)-bound bleomycin complexed with the bleomycin-binding protein from bleomycin-producing Streptomyces verticillus.
J.Biol.Chem., 277, 2002
|
|
3AHA
 
 | | Crystal structure of the complex between gp41 fragments N36 and C34 mutant N126K/E137Q | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Transmembrane protein gp41 | | 著者 | Izumi, K, Nakamura, S, Nakano, H, Shimura, K, Sakagami, Y, Oishi, S, Uchiyama, S, Ohkubo, T, Kobayashi, Y, Fujii, N, Matsuoka, M, Kodama, E.N. | | 登録日 | 2010-04-22 | | 公開日 | 2010-05-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Characterization of HIV-1 resistance to a fusion inhibitor, N36, derived from the gp41 amino terminal heptad repeat.
Antiviral Res., 2010
|
|
1ISE
 
 | | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly | | 分子名称: | Ribosome Recycling Factor | | 著者 | Nakano, H, Yoshida, T, Oka, S, Uchiyama, S, Nishina, K, Ohkubo, T, Kato, H, Yamagata, Y, Kobayashi, Y. | | 登録日 | 2001-11-30 | | 公開日 | 2003-10-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly
To be Published
|
|
