1VA2
 
 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 2) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1VA3
 
 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 3) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1VA1
 
 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 1) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1T7H
 
 | | X-ray structure of [Lys(-2)-Arg(-1)-des(17-21)]-endothelin-1 peptide | | Descriptor: | Endothelin-1 | | Authors: | Hoh, F, Cerdan, R, Kaas, Q, Nishi, Y, Chiche, L, Kubo, S, Chino, N, Kobayashi, Y, Dumas, C, Aumelas, A. | | Deposit date: | 2004-05-10 | | Release date: | 2004-12-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | High-resolution X-ray structure of the unexpectedly stable dimer of the [Lys(-2)-Arg(-1)-des(17-21)]endothelin-1 peptide
Biochemistry, 43, 2004
|
|
3AQC
 
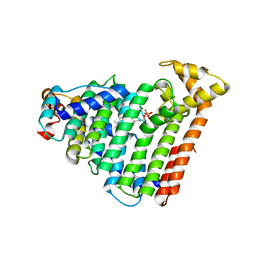 | | M. luteus B-P 26 heterodimeric hexaprenyl diphosphate synthase in complex with magnesium and FPP analogue | | Descriptor: | (2E,6E)-7,11-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, CHLORIDE ION, Component A of hexaprenyl diphosphate synthase, ... | | Authors: | Sasaki, D, Fujihashi, M, Okuyama, N, Kobayashi, Y, Noike, M, Koyama, T, Miki, K. | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of heterodimeric hexaprenyl diphosphate synthase from Micrococcus luteus B-P 26 reveals that the small subunit is directly involved in the product chain length regulation.
J.Biol.Chem., 286, 2011
|
|
3AQB
 
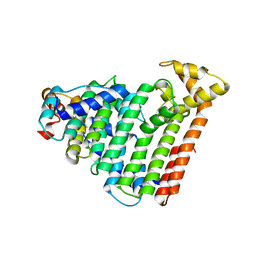 | | M. luteus B-P 26 heterodimeric hexaprenyl diphosphate synthase in complex with magnesium | | Descriptor: | CHLORIDE ION, Component A of hexaprenyl diphosphate synthase, Component B of hexaprenyl diphosphate synthase, ... | | Authors: | Sasaki, D, Fujihashi, M, Okuyama, N, Kobayashi, Y, Noike, M, Koyama, T, Miki, K. | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of heterodimeric hexaprenyl diphosphate synthase from Micrococcus luteus B-P 26 reveals that the small subunit is directly involved in the product chain length regulation.
J.Biol.Chem., 286, 2011
|
|
2KFJ
 
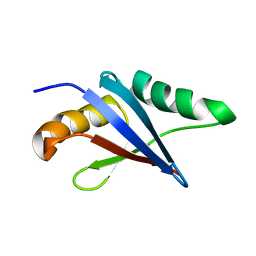 | | Solution structure of the loop deletion mutant of PB1 domain of Cdc24p | | Descriptor: | Cell division control protein 24 | | Authors: | Ogura, K, Tandai, T, Yoshinaga, S, Kobashigawa, Y, Kumeta, H, Inagaki, F. | | Deposit date: | 2009-02-22 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the heterodimer of Bem1 and Cdc24 PB1 domains from Saccharomyces cerevisiae
J.Biochem., 146, 2009
|
|
1VFH
 
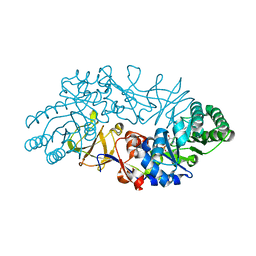 | | Crystal structure of alanine racemase from D-cycloserine producing Streptomyces lavendulae | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, alanine racemase | | Authors: | Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2004-04-13 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
J.Biol.Chem., 279, 2004
|
|
1VFS
 
 | | Crystal structure of D-cycloserine-bound form of alanine racemase from D-cycloserine-producing Streptomyces lavendulae | | Descriptor: | CHLORIDE ION, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, alanine racemase | | Authors: | Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2004-04-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
J.Biol.Chem., 279, 2004
|
|
2KE4
 
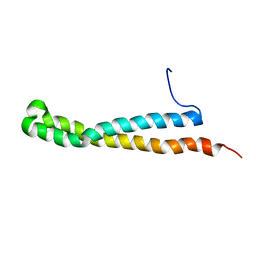 | |
2KBT
 
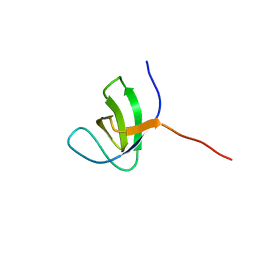 | | Attachment of an NMR-invisible solubility enhancement tag (INSET) using a sortase-mediated protein ligation method | | Descriptor: | Proto-oncogene vav,Immunoglobulin G-binding protein G | | Authors: | Kumeta, H, Kobashigawa, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2008-12-07 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Attachment of an NMR-invisible solubility enhancement tag using a sortase-mediated protein ligation method
J.Biomol.Nmr, 43, 2009
|
|
1WZB
 
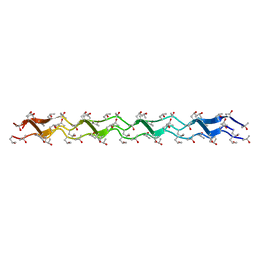 | | Crystal structure of the collagen triple helix model [{HYP(R)-HYP(R)-GLY}10]3 | | Descriptor: | Collagen triple helix | | Authors: | Kawahara, K, Nakamura, S, Nishi, Y, Uchiyama, S, Nishiuchi, Y, Nakazawa, T, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2005-03-03 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effect of hydration on the stability of the collagen-like triple-helical structure of [4(R)-hydroxyprolyl-4(R)-hydroxyprolylglycine]10
Biochemistry, 44, 2005
|
|
1EWJ
 
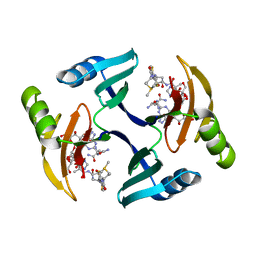 | | CRYSTAL STRUCTURE OF BLEOMYCIN-BINDING PROTEIN COMPLEXED WITH BLEOMYCIN | | Descriptor: | BLEOMYCIN A2, BLEOMYCIN RESISTANCE DETERMINANT | | Authors: | Maruyama, M, Kumagai, T, Matoba, Y, Hata, Y, Sugiyama, M. | | Deposit date: | 2000-04-26 | | Release date: | 2001-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the transposon Tn5-carried bleomycin resistance determinant uncomplexed and complexed with bleomycin.
J.Biol.Chem., 276, 2001
|
|
1DVV
 
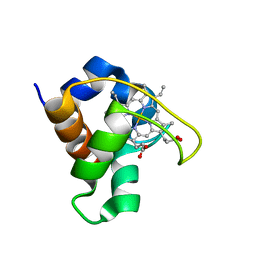 | | SOLUTION STRUCTURE OF THE QUINTUPLE MUTANT OF CYTOCHROME C-551 FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | CYTOCHROME C551, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hasegawa, J, Uchiyama, S, Tanimoto, Y, Mizutani, M, Kobayashi, Y, Sambongi, Y, Igarashi, Y. | | Deposit date: | 2000-01-22 | | Release date: | 2000-11-29 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Selected mutations in a mesophilic cytochrome c confer the stability of a thermophilic counterpart.
J.Biol.Chem., 275, 2000
|
|
1ECS
 
 | | THE 1.7 A CRYSTAL STRUCTURE OF A BLEOMYCIN RESISTANCE DETERMINANT ENCODED ON THE TRANSPOSON TN5 | | Descriptor: | BLEOMYCIN RESISTANCE PROTEIN, CALCIUM ION, TETRAETHYLENE GLYCOL | | Authors: | Maruyama, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2000-01-25 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the transposon Tn5-carried bleomycin resistance determinant uncomplexed and complexed with bleomycin.
J.Biol.Chem., 276, 2001
|
|
1WPK
 
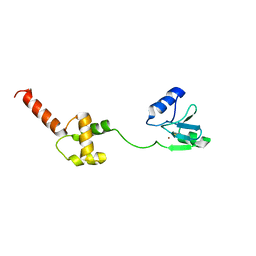 | | Methylated Form of N-terminal Transcriptional Regulator Domain of Escherichia Coli Ada Protein | | Descriptor: | ADA regulatory protein, ZINC ION | | Authors: | Takinowaki, H, Matsuda, Y, Yoshida, T, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2004-09-07 | | Release date: | 2005-09-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the methylated form of the N-terminal 16-kDa domain of Escherichia coli Ada protein
Protein Sci., 15, 2006
|
|
1GE9
 
 | | SOLUTION STRUCTURE OF THE RIBOSOME RECYCLING FACTOR | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Yoshida, T, Uchiyama, S, Nakano, H, Kashimori, H, Kijima, H, Ohshima, T, Saihara, Y, Ishino, T, Shimahara, T, Yoshida, T, Yokose, K, Ohkubo, T, Kaji, A, Kobayashi, Y. | | Deposit date: | 2000-10-19 | | Release date: | 2001-05-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosome recycling factor from Aquifex aeolicus.
Biochemistry, 40, 2001
|
|
1Y1C
 
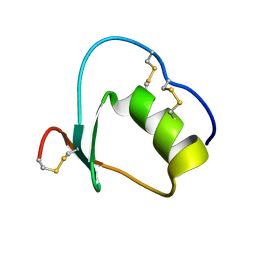 | | Solution structure of Anemonia elastase inhibitor analogue | | Descriptor: | Elastase inhibitor | | Authors: | Hemmi, H, Kumazaki, T, Yoshizawa-Kumagaye, K, Nishiuchi, Y, Yoshida, T, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2004-11-18 | | Release date: | 2005-07-19 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Study of an Anemonia Elastase Inhibitor, a "Nonclassical" Kazal-Type Inhibitor from Anemonia sulcata
Biochemistry, 44, 2005
|
|
1Y1B
 
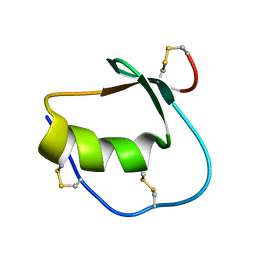 | | Solution structure of Anemonia elastase inhibitor | | Descriptor: | Elastase inhibitor | | Authors: | Hemmi, H, Kumazaki, T, Yoshizawa-Kumagaye, K, Nishiuchi, Y, Yoshida, T, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2004-11-18 | | Release date: | 2005-07-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Study of an Anemonia Elastase Inhibitor, a "Nonclassical" Kazal-Type Inhibitor from Anemonia sulcata
Biochemistry, 44, 2005
|
|
1V6R
 
 | | Solution Structure of Endothelin-1 with its C-terminal Folding | | Descriptor: | Endothelin-1 | | Authors: | Takashima, H, Mimura, N, Ohkubo, T, Yoshida, T, Tamaoki, H, Kobayashi, Y. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Distributed Computing and NMR Constraint-Based High-Resolution Structure
Determination: Applied for Bioactive Peptide Endothelin-1 To Determine C-Terminal
Folding
J.Am.Chem.Soc., 126, 2004
|
|
2LPU
 
 | | Solution structures of KmAtg10 | | Descriptor: | KmAtg10 | | Authors: | Yamaguchi, M, Noda, N.N, Yamamoto, H, Shima, T, Kumeta, H, Kobashigawa, Y, Akada, R, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-02-19 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into atg10-mediated formation of the autophagy-essential atg12-atg5 conjugate
Structure, 20, 2012
|
|
1WKI
 
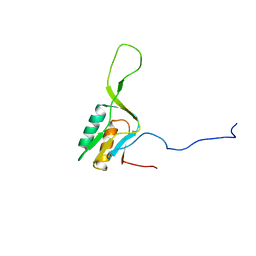 | | solution structure of ribosomal protein L16 from thermus thermophilus HB8 | | Descriptor: | LSU ribosomal protein L16P | | Authors: | Nishimura, M, Yoshida, T, Shirouzu, M, Terada, T, Kuramitsu, S, Yokoyama, S, Ohkubo, T, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ribosomal Protein L16 from Thermus thermophilus HB8
J.Mol.Biol., 344, 2004
|
|
1Y69
 
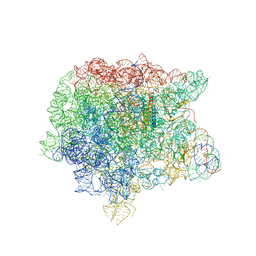 | | RRF domain I in complex with the 50S ribosomal subunit from Deinococcus radiodurans | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L16, 50S ribosomal protein L27, ... | | Authors: | Wilson, D.N, Schluenzen, F, Harms, J.M, Yoshida, T, Ohkubo, T, Albrecht, R, Buerger, J, Kobayashi, Y, Fucini, P. | | Deposit date: | 2004-12-04 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | X-ray crystallography on ribosome recycling: mechanism of binding and action of RRF on the 50S ribosomal subunit
EMBO J., 24, 2005
|
|
2LHI
 
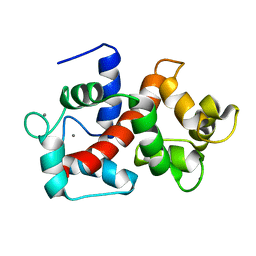 | | Solution structure of Ca2+/CNA1 peptide-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin,Serine/threonine-protein phosphatase 2B catalytic subunit A1 | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
2LHH
 
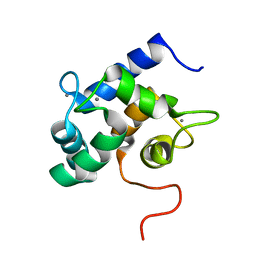 | | Solution structure of Ca2+-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
