3WAH
 
 | | Crystal Structure of HasAp with Iron MesoporphyrinIX | | Descriptor: | Heme acquisition protein HasAp, Mesoheme | | Authors: | Shirataki, C, Shoji, O, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2013-05-02 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Inhibition of Heme Uptake in Pseudomonas aeruginosa by its Hemophore (HasAp ) Bound to Synthetic Metal Complexes
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3WAX
 
 | | Crystal Structure of Autotaxin in Complex with 3BoA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Screening and X-ray Crystal Structure-based Optimization of Autotaxin (ENPP2) Inhibitors, Using a Newly Developed Fluorescence Probe
Acs Chem.Biol., 8, 2013
|
|
3WAY
 
 | | Crystal Structure of Autotaxin in Complex with 4BoA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Screening and X-ray Crystal Structure-based Optimization of Autotaxin (ENPP2) Inhibitors, Using a Newly Developed Fluorescence Probe
Acs Chem.Biol., 8, 2013
|
|
3WAV
 
 | | Crystal Structure of Autotaxin in Complex with Compound 10 | | Descriptor: | (5Z)-5-(3,4-dichlorobenzylidene)-2-(4-methylpiperazin-1-yl)-1,3-thiazol-4(5H)-one, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Screening and X-ray Crystal Structure-based Optimization of Autotaxin (ENPP2) Inhibitors, Using a Newly Developed Fluorescence Probe
Acs Chem.Biol., 8, 2013
|
|
3X2W
 
 | | Michaelis complex of cAMP-dependent Protein Kinase Catalytic Subunit | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, MAGNESIUM ION, ... | | Authors: | Das, A, Langan, P, Gerlits, O, Kovalevsky, A.Y, Heller, W.T. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein Kinase A Catalytic Subunit Primed for Action: Time-Lapse Crystallography of Michaelis Complex Formation.
Structure, 23, 2015
|
|
3X3B
 
 | | Crystal structure of the light-driven sodium pump KR2 in acidic state | | Descriptor: | DI(HYDROXYETHYL)ETHER, OLEIC ACID, RETINAL, ... | | Authors: | Kato, H.E, Inoue, K, Abe-Yoshizumi, R, Kato, Y, Ono, H, Konno, M, Ishizuka, T, Hoque, M.R, Hososhima, S, Kunitomo, H, Ito, J, Yoshizawa, S, Yamashita, K, Takemoto, M, Nishizawa, T, Taniguchi, R, Kogure, K, Maturana, A.D, Iino, Y, Yawo, H, Ishitani, R, Kandori, H, Nureki, O. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Na(+) transport mechanism by a light-driven Na(+) pump
Nature, 521, 2015
|
|
2F8B
 
 | |
6YAI
 
 | | Clathrin with bound beta2 appendage of AP2 | | Descriptor: | AP-2 complex subunit beta, Clathrin heavy chain, Clathrin light chain | | Authors: | Kovtun, O, Kane Dickson, V, Kelly, B.T, Owen, D, Briggs, J.A.G. | | Deposit date: | 2020-03-12 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Architecture of the AP2/clathrin coat on the membranes of clathrin-coated vesicles.
Sci Adv, 6, 2020
|
|
6YLN
 
 | | mTurquoise2 SG P212121 - Directional optical properties of fluorescent proteins | | Descriptor: | EGFP, POTASSIUM ION | | Authors: | Myskova, J, Rybakova, O, Brynda, J, Lazar, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y71
 
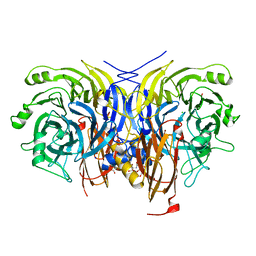 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H130A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
6YEY
 
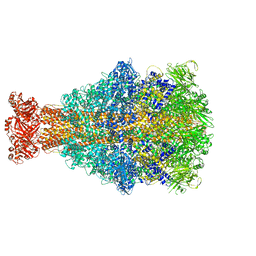 | | Xenorhabdus nematophila XptA1 in complex with porcine mucosa heparin | | Descriptor: | A component of insecticidal toxin complex (Tc) | | Authors: | Roderer, D, Broecker, F, Sitsel, O, Kaplonek, P, Leidreiter, F, Seeberger, P.H, Raunser, S. | | Deposit date: | 2020-03-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Glycan-dependent cell adhesion mechanism of Tc toxins.
Nat Commun, 11, 2020
|
|
6YP6
 
 | |
2GVS
 
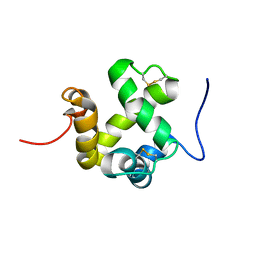 | | NMR solution structure of CSPsg4 | | Descriptor: | chemosensory protein CSP-sg4 | | Authors: | Tomaselli, S, Crescenzi, O, Sanfelice, D, Ab, E, Tancredi, T, Picone, D. | | Deposit date: | 2006-05-03 | | Release date: | 2006-09-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Chemosensory Protein from the Desert Locust Schistocerca gregaria(,).
Biochemistry, 45, 2006
|
|
6YD9
 
 | | Ecoli GyrB24 with inhibitor 16a | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, N-[6-(3-azanylpropanoylamino)-1,3-benzothiazol-2-yl]-3,4-bis(chloranyl)-5-methyl-1H-pyrrole-2-carboxamide | | Authors: | Barancokova, M, Skok, Z, Benek, O, Cruz, C.D, Tammela, P, Tomasic, T, Zidar, N, Masic, L.P, Zega, A, Stevenson, C.E.M, Mundy, J, Lawson, D.M, Maxwell, A.M, Kikelj, D, Ilas, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploring the Chemical Space of Benzothiazole-Based DNA Gyrase B Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6Y2N
 
 | | Crystal structure of ribonucleotide reductase R2 subunit solved by serial synchrotron crystallography | | Descriptor: | FE (III) ION, MANGANESE (III) ION, Ribonucleoside-diphosphate reductase subunit beta | | Authors: | Shilova, A, Lebrette, H, Aurelius, O, Hogbom, M, Mueller, U. | | Deposit date: | 2020-02-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Current status and future opportunities for serial crystallography at MAX IV Laboratory.
J.Synchrotron Radiat., 27, 2020
|
|
6YAE
 
 | | AP2 core in physiological buffer | | Descriptor: | AP-2 complex subunit alpha, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Kane Dickson, V, Kovtun, O, Kelly, B.T, Owen, D.J, Briggs, J.A.G. | | Deposit date: | 2020-03-12 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the AP2/clathrin coat on the membranes of clathrin-coated vesicles.
Sci Adv, 6, 2020
|
|
6YGB
 
 | | Crystal structure of the NatC complex bound to CoA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Grunwald, S, Hopf, L, Bock-Bierbaum, T, Lally, C.C, Spahn, C.M.T, Daumke, O. | | Deposit date: | 2020-03-27 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Divergent architecture of the heterotrimeric NatC complex explains N-terminal acetylation of cognate substrates.
Nat Commun, 11, 2020
|
|
6Y6Y
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H129A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
6Y7E
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H494A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
6YLO
 
 | | mTurquoise2 - Directionality of Optical Properties of Fluorescent Proteins | | Descriptor: | POTASSIUM ION, TETRAETHYLENE GLYCOL, mTurquoise2_C2221 | | Authors: | Myskova, J, Rybakova, O, Brynda, J, Lazar, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YLS
 
 | | mEos4b - Directionality of Optical Properties of Fluorescent Proteins | | Descriptor: | CHLORIDE ION, Green to red photoconvertible GFP-like protein EosFP, SODIUM ION | | Authors: | Myskova, J, Rybakova, O, Brynda, J, Lazar, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y2L
 
 | | Structure of human ribosome in POST state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-16 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
6Y7D
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H433A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
2HKU
 
 | | Structural Genomics, the crystal structure of a putative transcriptional regulator from Rhodococcus sp. RHA1 | | Descriptor: | 1,2-ETHANEDIOL, A putative transcriptional regulator, TETRAETHYLENE GLYCOL | | Authors: | Tan, K, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a transcriptional regulator from Rhodococcus sp. RHA1
To be Published
|
|
6YAH
 
 | | AP2 in clathrin coats assembled on a membrane containing dileucine- and tyrosine-based cargo peptides | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Kovtun, O, Kane Dickson, V, Kelly, B.T, Owen, D, Briggs, J.A.G. | | Deposit date: | 2020-03-12 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (10.2 Å) | | Cite: | Architecture of the AP2/clathrin coat on the membranes of clathrin-coated vesicles.
Sci Adv, 6, 2020
|
|
