1TZ4
 
 | | [hPP19-23]-pNPY bound to DPC Micelles | | Descriptor: | neuropeptide Y,Pancreatic prohormone,neuropeptide Y | | Authors: | Lerch, M, Kamimori, H, Folkers, G, Aguilar, M.I, Beck-Sickinger, A.G, Zerbe, O. | | Deposit date: | 2004-07-09 | | Release date: | 2005-07-05 | | Last modified: | 2020-01-01 | | Method: | SOLUTION NMR | | Cite: | Strongly Altered Receptor Binding Properties in PP and NPY Chimeras Are Accompanied by Changes
in Structure and Membrane Binding
Biochemistry, 44, 2005
|
|
7R0R
 
 | |
4UN3
 
 | | Crystal structure of Cas9 bound to PAM-containing DNA target | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
1UBU
 
 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
7RAN
 
 | | 5-HT2AR bound to a novel agonist in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | (3R)-3-methyl-5-(1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,3,6-tetrahydropyridin-1-ium, 5-hydroxytryptamine receptor 2A, G protein subunit q (Gi2-mini-Gq chimera), ... | | Authors: | Barros-Alvarez, X, Kim, K, Panova, O, Roth, B.L, Skiniotis, G. | | Deposit date: | 2021-07-02 | | Release date: | 2022-07-06 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Bespoke library docking for 5-HT 2A receptor agonists with antidepressant activity.
Nature, 610, 2022
|
|
4UPA
 
 | |
1SEG
 
 | | Crystal structure of a toxin chimera between Lqh-alpha-IT from the scorpion Leiurus quinquestriatus hebraeus and AAH2 from Androctonus australis hector | | Descriptor: | AAH2: LQH-ALPHA-IT (FACE) CHIMERIC TOXIN, NITRATE ION, PROPANOIC ACID, ... | | Authors: | Karbat, I, Frolow, F, Froy, O, Gilles, N, Cohen, L, Turkov, M, Gordon, D, Gurevitz, M. | | Deposit date: | 2004-02-17 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis of the high insecticidal potency of scorpion alpha-toxins.
J.Biol.Chem., 279, 2004
|
|
4U7K
 
 | | Crystal structure of the Clostridium histolyticum colH collagenase polycystic kidney disease-like domain 2a in the presence of calcium at 1.9 Angstrom resolution | | Descriptor: | CALCIUM ION, ColH protein | | Authors: | Bauer, R, Janowska, K, Sakon, J, Matsushita, O, Latimer, E. | | Deposit date: | 2014-07-31 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UP7
 
 | |
7RKB
 
 | | Crystal Structure of Putative Pterin Binding Protein (PruR) from Klebsiella pneumoniae in Complex with Neopterin | | Descriptor: | CHLORIDE ION, L-NEOPTERIN, Pterin Binding Protein, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-22 | | Release date: | 2022-08-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Putative Pterin Binding Protein (PruR) from Klebsiella pneumoniae in Complex with Neopterin.
To Be Published
|
|
7RNX
 
 | | CLC-ec1 at pH 7.5 100mM Cl Swap | | Descriptor: | H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Boudker, O, Accardi, A. | | Deposit date: | 2021-07-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural basis of common gate activation in CLC transporters
To Be Published
|
|
7RP6
 
 | |
1SL5
 
 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with LNFP III (Dextra L504). | | Descriptor: | CALCIUM ION, MAGNESIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
7RO0
 
 | |
4UE9
 
 | |
7RSB
 
 | |
7RP5
 
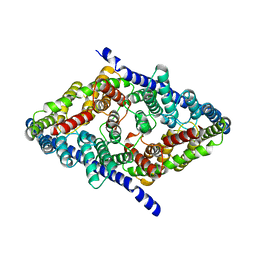 | | CLC-ec1 at pH 4.5 100mM Cl Turn | | Descriptor: | H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Boudker, O, Accardi, A. | | Deposit date: | 2021-08-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis of common gate activation in CLC transporters
To Be Published
|
|
1RQM
 
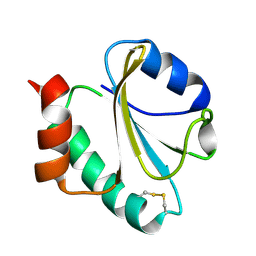 | | SOLUTION STRUCTURE OF THE K18G/R82E ALICYCLOBACILLUS ACIDOCALDARIUS THIOREDOXIN MUTANT | | Descriptor: | Thioredoxin | | Authors: | Leone, M, Di Lello, P, Ohlenschlager, O, Pedone, E.M, Bartolucci, S, Rossi, M, Di Blasio, B, Pedone, C, Saviano, M, Isernia, C, Fattorusso, R. | | Deposit date: | 2003-12-05 | | Release date: | 2004-06-22 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the K18G/R82E Alicyclobacillus acidocaldarius Thioredoxin Mutant: A Molecular Analysis of Its Reduced Thermal Stability.
Biochemistry, 43, 2004
|
|
7RQ7
 
 | |
7R6S
 
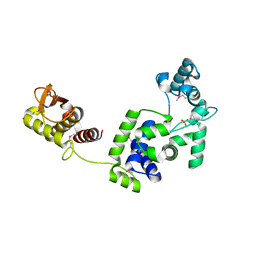 | | Crystal Structure of the Putative Bacteriophage Protein from Stenotrophomonas maltophilia | | Descriptor: | Putative bacteriophage protein, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Brunzelle, J.S, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2021-06-23 | | Release date: | 2022-11-09 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Putative Bacteriophage Protein from Stenotrophomonas maltophilia
To Be Published
|
|
1R1X
 
 | | Crystal structure of oxy-human hemoglobin Bassett at 2.15 angstrom | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Abdulmalik, O, Safo, M.K, Lerner, N.B, Ochotorena, J, Dhaikin, Y, Abraham, D.J, Asakura, T. | | Deposit date: | 2003-09-25 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Characterization of hemoglobin bassett (alpha94Asp-->Ala), a variant with very low oxygen affinity
Am.J.Hematol., 77, 2004
|
|
4UP8
 
 | |
4UN5
 
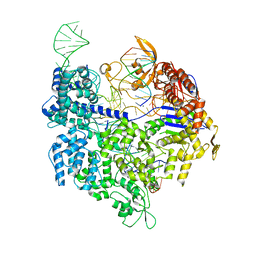 | | Crystal structure of Cas9 bound to PAM-containing DNA target containing mismatches at positions 1-3 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
4UN4
 
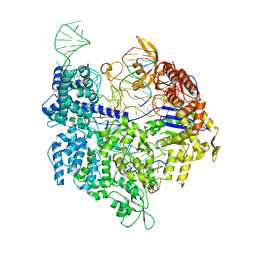 | | Crystal structure of Cas9 bound to PAM-containing DNA target with mismatches at positions 1-2 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.371 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
4UMV
 
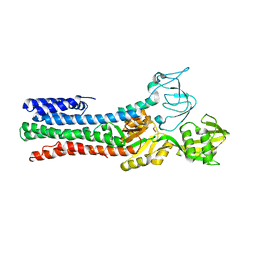 | | CRYSTAL STRUCTURE OF A ZINC-TRANSPORTING PIB-TYPE ATPASE IN THE E2P STATE | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ZINC-TRANSPORTING ATPASE | | Authors: | Wang, K.T, Sitsel, O, Meloni, G, Autzen, H.E, Andersson, M, Klymchuk, T, Nielsen, A.M, Rees, D.C, Nissen, P, Gourdon, P. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Mechanism of Zn(2+)-Transporting P-Type Atpases.
Nature, 514, 2014
|
|
