7COL
 
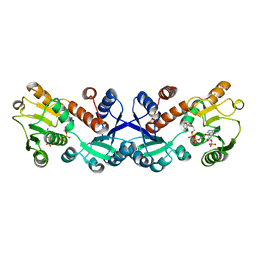 | | Crystal structure of 5-ketofructose reductase complexed with NADPH | | Descriptor: | 5-ketofructose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hodoya, Y, Noda, S, Nguyen, T.M, Kataoka, N, Adachi, O, Matsutani, M, Matsushita, K, Yakushi, T, Goto, M. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The 5-Ketofructose Reductase of Gluconobacter sp. Strain CHM43 Is a Novel Class in the Shikimate Dehydrogenase Family.
J.Bacteriol., 203, 2021
|
|
7CVR
 
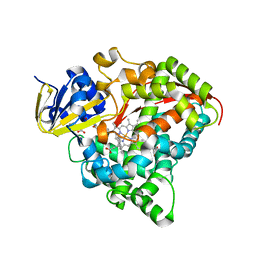 | | Structure of the CYP102A1 Haem Domain with N-Carboxybenzyl-L-Prolyl-L-Phenylalanine in complex with (S)-1-Tetralylamine | | Descriptor: | (1~{S})-1,2,3,4-tetrahydronaphthalen-1-amine, (2S)-3-phenyl-2-[[(2S)-1-phenylmethoxycarbonylpyrrolidin-2-yl]carbonylamino]propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Stanfield, J.K, Sugimoto, H, Shoji, O. | | Deposit date: | 2020-08-26 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the CYP102A1 Haem Domain with N-Carboxybenzyl-L-Prolyl-L-Phenylalanine in complex with (S)-1-Tetralylamine at 1.60 Angstrom Resolution
To Be Published
|
|
7CX6
 
 | |
7CX8
 
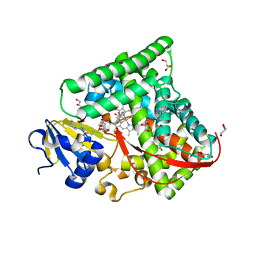 | | Structure of the CYP102A1 Haem Domain with N-(5-Cyclohexyl)valeroyl-L-Phenylalanine in complex with (R)-1-Tetralylamine | | Descriptor: | (1R)-1,2,3,4-tetrahydronaphthalen-1-amine, (1~{S})-1,2,3,4-tetrahydronaphthalen-1-amine, (2~{S})-2-(5-cyclohexylpentanoylamino)-3-phenyl-propanoic acid, ... | | Authors: | Stanfield, J.K, Sugimoto, H, Shoji, O. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the CYP102A1 Haem Domain with N-(5-Cyclohexyl)valeroyl-L-Phenylalanine in complex with (R)-1-Tetralylamine at 1.70 Angstrom Resolution
To Be Published
|
|
7CKN
 
 | |
7COO
 
 | |
7CON
 
 | |
7CP8
 
 | |
7D3S
 
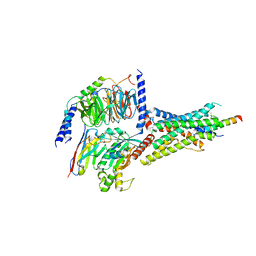 | | Human SECR in complex with an engineered Gs heterotrimer | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Fukuhara, S, Kobayashi, K, Kusakizako, T, Shihoya, W, Nureki, O. | | Deposit date: | 2020-09-20 | | Release date: | 2020-11-04 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the human secretin receptor coupled to an engineered heterotrimeric G protein.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7D7P
 
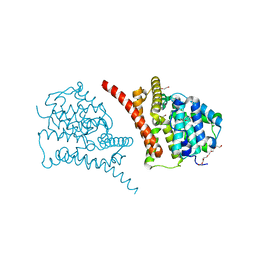 | | Crystal structure of the phosphodiesterase domain of Salpingoeca rosetta rhodopsin phosphodiesterase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Ikuta, T, Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2020-10-05 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the mechanism of rhodopsin phosphodiesterase.
Nat Commun, 11, 2020
|
|
7D16
 
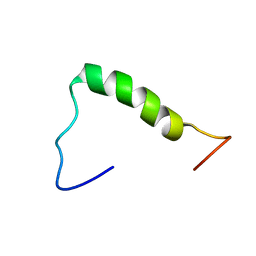 | | NERP-2 in a DPC solution | | Descriptor: | Neurosecretory protein VGF | | Authors: | Park, O, Cheong, C, Jeon, Y. | | Deposit date: | 2020-09-13 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of neuroendocrine regulatory peptide-2 in membrane-mimicking environments.
Peptide Science, 2020
|
|
3PCQ
 
 | | Femtosecond X-ray protein Nanocrystallography | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chapman, H.N, Fromme, P, Barty, A, White, T.A, Kirian, R.A, Aquila, A, Hunter, M.S, Schulz, J, Deponte, D.P, Weierstall, U, Doak, R.B, Maia, F.R.N.C, Martin, A.V, Schlichting, I, Lomb, L, Coppola, N, Shoeman, R.L, Epp, S.W, Hartmann, R, Rolles, D, Rudenko, A, Foucar, L, Kimmel, N, Weidenspointner, G, Holl, P, Liang, M, Barthelmess, M, Caleman, C, Boutet, S, Bogan, M.J, Krzywinski, J, Bostedt, C, Bajt, S, Gumprecht, L, Rudek, B, Erk, B, Schmidt, C, Homke, A, Reich, C, Pietschner, D, Struder, L, Hauser, G, Gorke, H, Ullrich, J, Herrmann, S, Schaller, G, Schopper, F, Soltau, H, Kuhnel, K.-U, Messerschmidt, M, Bozek, J.D, Hau-Riege, S.P, Frank, M, Hampton, C.Y, Sierra, R, Starodub, D, Williams, G.J, Hajdu, J, Timneanu, N, Seibert, M.M, Andreasson, J, Rocker, A, Jonsson, O, Svenda, M, Stern, S, Nass, K, Andritschke, R, Schroter, C.-D, Krasniqi, F, Bott, M, Schmidt, K.E, Wang, X, Grotjohann, I, Holton, J.M, Barends, T.R.M, Neutze, R, Marchesini, S, Fromme, R, Schorb, S, Rupp, D, Adolph, M, Gorkhover, T, Andersson, I, Hirsemann, H, Potdevin, G, Graafsma, H, Nilsson, B, Spence, J.C.H. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (8.984 Å) | | Cite: | Femtosecond X-ray protein nanocrystallography.
Nature, 470, 2011
|
|
7DH5
 
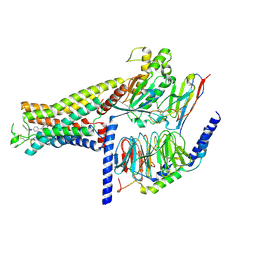 | | Dog beta3 adrenergic receptor bound to mirabegron in complex with a miniGs heterotrimer | | Descriptor: | 2-(2-azanyl-1,3-thiazol-4-yl)-N-[4-[2-[[(2R)-2-oxidanyl-2-phenyl-ethyl]amino]ethyl]phenyl]ethanamide, Beta-3 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2020-11-12 | | Release date: | 2021-08-04 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structure of the beta 3-adrenergic receptor reveals the molecular basis of subtype selectivity.
Mol.Cell, 81, 2021
|
|
7DB6
 
 | | human melatonin receptor MT1 - Gi1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Okamoto, H.H, Kusakizako, T, Shihioya, W, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-10-19 | | Release date: | 2021-08-18 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the human MT 1 -G i signaling complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2MXS
 
 | | Solution NMR-structure of the neomycin sensing riboswitch RNA bound to paromomycin | | Descriptor: | PAROMOMYCIN, RNA (27-MER) | | Authors: | Schmidtke, S, Duchardt-Ferner, E, Ohlenschlaeger, O, Gottstein, D, Wohnert, J. | | Deposit date: | 2015-01-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
7CZI
 
 | |
7D0T
 
 | |
7D0U
 
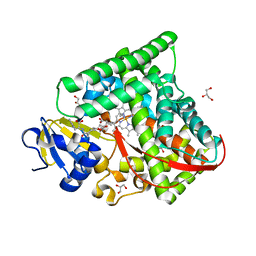 | |
7D1F
 
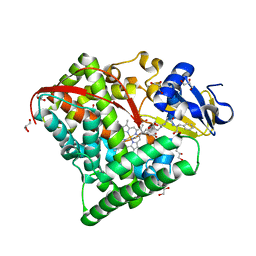 | |
6OV8
 
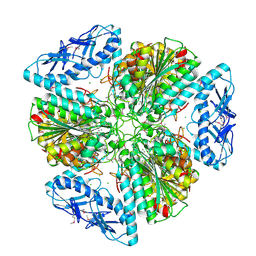 | | 2.6 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Peptidase B, ... | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-07 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
6OAD
 
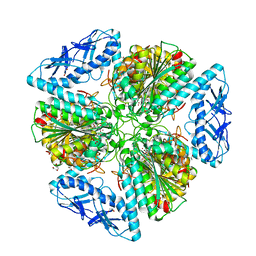 | | 2.05 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655. | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-03-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
7B7A
 
 | | ENDO-POLYGALACTURONASE FROM ARABIDOPSIS THALIANA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pectin lyase-like superfamily protein, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Safran, J, Tabi, W, Habrylo, O, Bouckaert, J, Lefebvre, V, Senechal, F, Pelloux, J. | | Deposit date: | 2020-12-10 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Plant polygalacturonase structures specify enzyme dynamics and processivities to fine-tune cell wall pectins.
Plant Cell, 2023
|
|
7B8B
 
 | | ADPG2 - ENDOPOLYGALACTURONASE FROM ARABIDOPSIS THALIANA | | Descriptor: | PHOSPHATE ION, Polygalacturonase ADPG2 | | Authors: | Safran, J, Tabi, W, Habrylo, O, Bouckaert, J, Lefebvre, V, Senechal, F, Pelloux, J. | | Deposit date: | 2020-12-12 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Plant polygalacturonase structures specify enzyme dynamics and processivities to fine-tune cell wall pectins.
Plant Cell, 2023
|
|
7D7Q
 
 | | Crystal structure of the transmembrane domain and linker region of Salpingoeca rosetta rhodopsin phosphodiesterase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phosphodiesterase, RETINAL | | Authors: | Ikuta, T, Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2020-10-05 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into the mechanism of rhodopsin phosphodiesterase.
Nat Commun, 11, 2020
|
|
7D13
 
 | | NERP-2 in a HFIP solution | | Descriptor: | Neurosecretory protein VGF | | Authors: | Park, O, Cheong, C, Jeon, Y. | | Deposit date: | 2020-09-13 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of neuroendocrine regulatory peptide-2 in membrane-mimicking environments.
Peptide Science, 2020
|
|
