8OUT
 
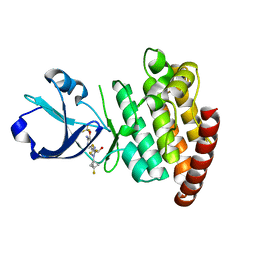 | | CRYSTAL STRUCTURE OF DLK IN COMPLEX WITH COMPOUND 22 | | 分子名称: | (1R)-1-[4-[6-azanyl-5-(trifluoromethyloxy)pyridin-3-yl]-1-(3-fluoranyl-1-bicyclo[1.1.1]pentanyl)imidazol-2-yl]-2,2,2-tris(fluoranyl)ethanol, Mitogen-activated protein kinase kinase kinase 12 | | 著者 | Zebisch, M, Akkermans, O, Barker, J.J, Cross, J.B. | | 登録日 | 2023-04-24 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.935 Å) | | 主引用文献 | Discovery of IACS-52825, a Potent and Selective DLK Inhibitor for Treatment of Chemotherapy-Induced Peripheral Neuropathy.
J.Med.Chem., 66, 2023
|
|
8OV0
 
 | | MUC5AC CysD7 amino acids 3518-3626 | | 分子名称: | CALCIUM ION, CHLORIDE ION, Mucin-5AC | | 著者 | Khmelnitsky, L, Milo, A, Dym, O, Fass, D. | | 登録日 | 2023-04-25 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Diversity of CysD domains in gel-forming mucins.
Febs J., 290, 2023
|
|
2E81
 
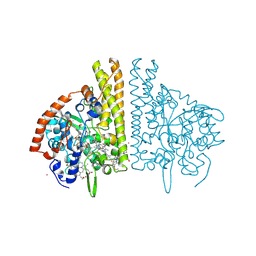 | |
2E80
 
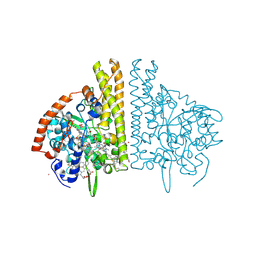 | |
3NPV
 
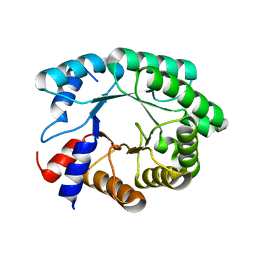 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution | | 分子名称: | deoxyribose phosphate aldolase | | 著者 | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2010-06-29 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
6GEL
 
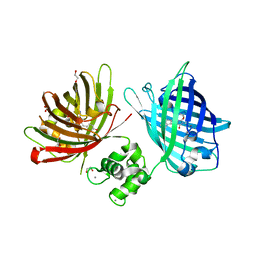 | | The structure of TWITCH-2B | | 分子名称: | CALCIUM ION, FORMIC ACID, GLYCEROL, ... | | 著者 | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | 登録日 | 2018-04-26 | | 公開日 | 2019-08-21 | | 最終更新日 | 2019-09-11 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
3NQ2
 
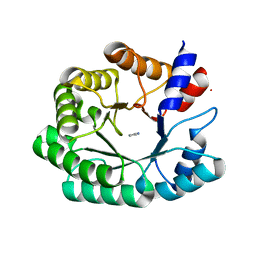 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution R2 3/5G | | 分子名称: | IMIDAZOLE, deoxyribose phosphate aldolase | | 著者 | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2010-06-29 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3NR0
 
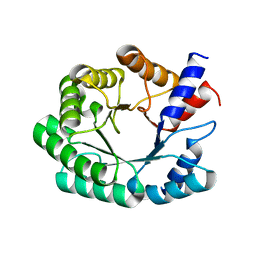 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution R6 6/10A | | 分子名称: | deoxyribose phosphate aldolase | | 著者 | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2010-06-30 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3NPX
 
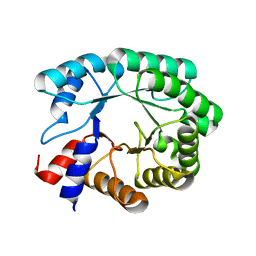 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution | | 分子名称: | deoxyribose phosphate aldolase | | 著者 | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2010-06-29 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
5N49
 
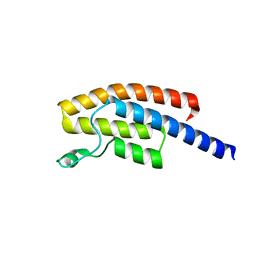 | | BRPF2 in complex with Compound 7 | | 分子名称: | 2-(1,3,6-trimethyl-2-oxidanylidene-benzimidazol-5-yl)benzo[de]isoquinoline-1,3-dione, Bromodomain-containing protein 1 | | 著者 | Bouche, L, Christ, C.D, Siegel, S, Fernandez-Montalvan, A.E, Holton, S.J, Fedorov, O, ter Laak, A, Sugawara, T, Stoeckigt, D, Tallant, C, Bennett, J, Monteiro, O, Saez, L.D, Siejka, P, Meier, J, Puetter, V, Weiske, J, Mueller, S, Huber, K.V.M, Hartung, I.V, Haendler, B. | | 登録日 | 2017-02-10 | | 公開日 | 2017-05-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains.
J. Med. Chem., 60, 2017
|
|
3TBJ
 
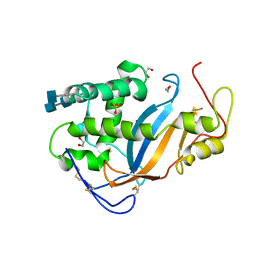 | |
3OOM
 
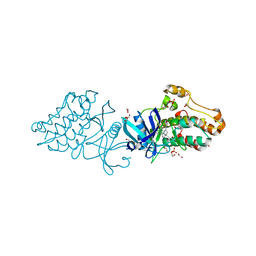 | | Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00507 | | 分子名称: | 1,2-ETHANEDIOL, 1-{3-[6-(tetrahydro-2H-pyran-4-ylamino)imidazo[1,2-b]pyridazin-3-yl]phenyl}ethanone, Activin receptor type-1, ... | | 著者 | Chaikuad, A, Sanvitale, C, Cooper, C, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Gileadi, O, Fedorov, O, Allerston, C, Krojer, T, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | 登録日 | 2010-08-31 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00507
To be Published
|
|
3OP5
 
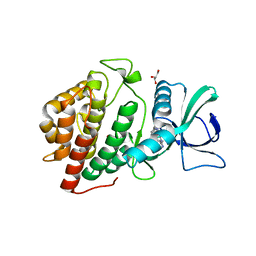 | | Human vaccinia-related kinase 1 | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Serine/threonine-protein kinase VRK1, ... | | 著者 | Allerston, C.K, Uttarkar, S, Savitsky, P, Elkins, J.M, Filippakopoulos, P, Krojer, T, Rellos, P, Fedorov, O, Eswaran, J, Brenner, B, Keates, T, Das, S, King, O, Chalk, R, Berridge, G, von Delft, F, Gileadi, O, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2010-08-31 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural characterization of human Vaccinia-Related Kinases (VRK) bound to small-molecule inhibitors identifies different P-loop conformations.
Sci Rep, 7, 2017
|
|
2UAG
 
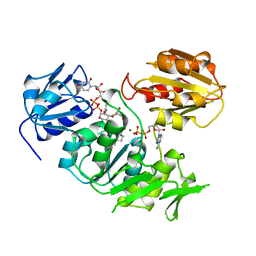 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE), ... | | 著者 | Bertrand, J, Fanchon, E, Dideberg, O. | | 登録日 | 1999-02-23 | | 公開日 | 2000-02-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Determination of the MurD mechanism through crystallographic analysis of enzyme complexes.
J.Mol.Biol., 289, 1999
|
|
3Q2D
 
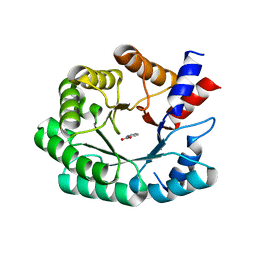 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution | | 分子名称: | 5-nitro-1H-benzotriazole, Deoxyribose phosphate aldolase | | 著者 | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Murphy, P, Dym, O, Albeck, S, Kiss, G, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2010-12-20 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3VCR
 
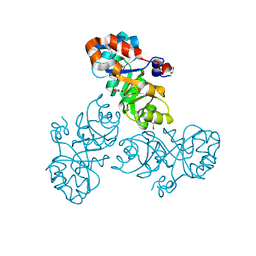 | | Crystal structure of a putative Kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase from Oleispira antarctica | | 分子名称: | PYRUVIC ACID, putative Kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase | | 著者 | Stogios, P.J, Kagan, O, Di Leo, R, Yim, V, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-01-04 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3SZR
 
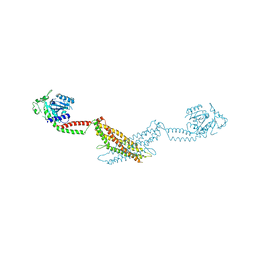 | |
2V1X
 
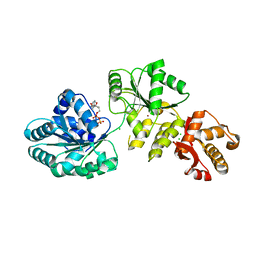 | | Crystal structure of human RECQ-like DNA helicase | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT DNA HELICASE Q1, ... | | 著者 | Pike, A.C.W, Shrestha, B, Burgess-Brown, N, King, O, Ugochukwu, E, Watt, S, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Gileadi, O. | | 登録日 | 2007-05-30 | | 公開日 | 2007-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the Human Recq1 Helicase Reveals a Putative Strand-Separation Pin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3U5K
 
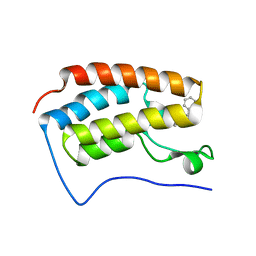 | | Crystal Structure of the first bromodomain of human BRD4 in complex with Midazolam | | 分子名称: | 8-chloro-6-(2-fluorophenyl)-1-methyl-4H-imidazo[1,5-a][1,4]benzodiazepine, Bromodomain-containing protein 4 | | 著者 | Filippakopoulos, P, Picaud, S, Felletar, I, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2011-10-11 | | 公開日 | 2011-11-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Benzodiazepines and benzotriazepines as protein interaction inhibitors targeting bromodomains of the BET family.
Bioorg.Med.Chem., 20, 2012
|
|
3NQV
 
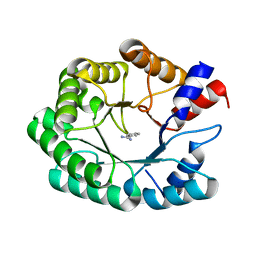 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution R5 7/4A | | 分子名称: | BENZAMIDINE, deoxyribose phosphate aldolase | | 著者 | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2010-06-29 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3UG9
 
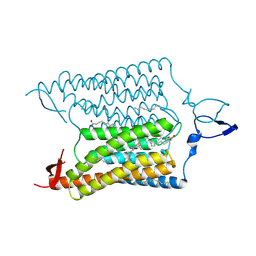 | | Crystal Structure of the Closed State of Channelrhodopsin | | 分子名称: | Archaeal-type opsin 1, Archaeal-type opsin 2, OLEIC ACID, ... | | 著者 | Kato, H.E, Ishitani, R, Nureki, O. | | 登録日 | 2011-11-02 | | 公開日 | 2012-01-25 | | 最終更新日 | 2017-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the channelrhodopsin light-gated cation channel
Nature, 482, 2012
|
|
6GEZ
 
 | | THE STRUCTURE OF TWITCH-2B N532F | | 分子名称: | CALCIUM ION, FORMIC ACID, Green fluorescent protein,Optimized Ratiometric Calcium Sensor,Green fluorescent protein,Green fluorescent protein | | 著者 | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | 登録日 | 2018-04-27 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
3V77
 
 | | Crystal structure of a putative fumarylacetoacetate isomerase/hydrolase from Oleispira antarctica | | 分子名称: | ACETATE ION, D(-)-TARTARIC ACID, Putative fumarylacetoacetate isomerase/hydrolase, ... | | 著者 | Stogios, P.J, Kagan, O, Di Leo, R, Bochkarev, A, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-12-20 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3NPU
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution | | 分子名称: | deoxyribose phosphate aldolase | | 著者 | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2010-06-29 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3NPW
 
 | | In silico designed of an improved Kemp eliminase KE70 mutant by computational design and directed evolution | | 分子名称: | deoxyribose phosphate aldolase | | 著者 | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2010-06-29 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
