8F4E
 
 | | RT XFEL structure of Photosystem II 250 microseconds after the third illumination at 2.09 Angstrom resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | 登録日 | 2022-11-10 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4G
 
 | | RT XFEL structure of Photosystem II 730 microseconds after the third illumination at 2.03 Angstrom resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | 登録日 | 2022-11-10 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4J
 
 | | RT XFEL structure of Photosystem II 4000 microseconds after the third illumination at 2.00 Angstrom resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | 登録日 | 2022-11-10 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4Y
 
 | | Crystal Structure of SARS-CoV-2 2'-O-Methyltransferase in Complex with Compound 5a covalently bound to nsp16 and nsp10 | | 分子名称: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, 4-[2-(2,4-dichlorophenyl)ethyl]-6-(trifluoromethyl)pyrimidin-2-ol, ... | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-11-11 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8F4S
 
 | | Crystal Structure of the SARS-CoV-2 2'-O-Methyltransferase with Compound 5a bound to the Cryptic Pocket of nsp16 | | 分子名称: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, FORMIC ACID, ... | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-11-11 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8GH3
 
 | | Structure of Trypanosoma (MDH)4-(Pex5)2, distal conformation | | 分子名称: | Peroxisome targeting signal 1 receptor, malate dehydrogenase | | 著者 | Sonani, R.R, Artur, B, Jemiola-Rzeminska, M, Lipinski, O, Patel, S.N, Sood, T, Dubin, G. | | 登録日 | 2023-03-09 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | Structure of Trypanosoma (MDH)4-(Pex5)2, distal conformation
To Be Published
|
|
8GH2
 
 | | Structure of Trypanosoma (MDH)4-(Pex5)2, close conformation | | 分子名称: | Peroxisome targeting signal 1 receptor, malate dehydrogenase | | 著者 | Sonani, R.R, Artur, B, Jemiola-Rzeminska, M, Lipinski, O, Patel, S.N, Sood, T, Dubin, G. | | 登録日 | 2023-03-09 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Structure of Trypanosoma (MDH)4-(Pex5)2, close conformation
To Be Published
|
|
8GI0
 
 | | Structure of Trypanosoma docking complex | | 分子名称: | Peroxisomal membrane protein PEX14, Peroxisome targeting signal 1 receptor, malate dehydrogenase | | 著者 | Sonani, R.R, Artur, B, Jemiola-Rzeminska, M, Lipinski, O, Patel, S.N, Sood, T, Dubin, G. | | 登録日 | 2023-03-13 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of Trypanosoma docking complex
To Be Published
|
|
8GGH
 
 | | Structure of Trypanosoma (MDH)4-PEX5, distal conformation | | 分子名称: | Peroxisome targeting signal 1 receptor, malate dehydrogenase | | 著者 | Sonani, R.R, Artur, B, Jemiola-Rzeminska, M, Lipinski, O, Patel, S.N, Sood, T, Dubin, G. | | 登録日 | 2023-03-08 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Structure of Trypanosoma (MDH)4-PEX5, distal conformation
To Be Published
|
|
8GGD
 
 | | Structure of Trypanosoma (MDH)4-Pex5, close conformation | | 分子名称: | Peroxisome targeting signal 1 receptor, malate dehydrogenase | | 著者 | Sonani, R.R, Artur, B, Jemiola-Rzeminska, M, Lipinski, O, Patel, S.N, Sood, T, Dubin, G. | | 登録日 | 2023-03-08 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Structure of Trypanosoma (MDH)4-Pex5, close conformation
To Be Published
|
|
1WQ3
 
 | | Escherichia coli tyrosyl-tRNA synthetase mutant complexed with 3-iodo-L-tyrosine | | 分子名称: | 3-IODO-TYROSINE, Tyrosyl-tRNA synthetase | | 著者 | Kobayashi, T, Sakamoto, K, Nureki, O, Takimura, T, Kamata, K, Sekine, R, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-09-20 | | 公開日 | 2005-01-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1WQ4
 
 | | Escherichia coli tyrosyl-tRNA synthetase mutant complexed with L-tyrosine | | 分子名称: | TYROSINE, Tyrosyl-tRNA synthetase | | 著者 | Kobayashi, T, Sakamoto, K, Nureki, O, Takimura, T, Kamata, K, Sekine, R, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-09-20 | | 公開日 | 2005-01-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6UHX
 
 | | Crystal structure of YIR035C short chain dehydrogenases/reductase from Saccharomyces cerevisiae | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized oxidoreductase YIR035C | | 著者 | Stogios, P.J, Skarina, T, Chen, C, Kagan, O, Iakounine, A, Savchenko, A. | | 登録日 | 2019-09-29 | | 公開日 | 2020-08-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal structure of YIR035C short chain dehydrogenases/reductase from Saccharomyces cerevisiae
To Be Published
|
|
6TEO
 
 | | Crystal structure of a yeast Snu114-Prp8 complex | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Pre-mRNA-splicing factor 8, ... | | 著者 | Ganichkin, O, Jia, J, Loll, B, Absmeier, E, Wahl, M.C. | | 登録日 | 2019-11-12 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | A Snu114-GTP-Prp8 module forms a relay station for efficient splicing in yeast.
Nucleic Acids Res., 48, 2020
|
|
1YDX
 
 | | Crystal structure of Type-I restriction-modification system S subunit from M. genitalium | | 分子名称: | CHLORIDE ION, type I restriction enzyme specificity protein MG438 | | 著者 | Machado, B, Quijada, O, Pinol, J, Fita, I, Querol, E, Carpena, X. | | 登録日 | 2004-12-27 | | 公開日 | 2005-08-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of a Putative Type I Restriction-Modification S Subunit from Mycoplasma genitalium
J.Mol.Biol., 351, 2005
|
|
6TLY
 
 | | Crystal structure of the unconventional kinetochore protein Bodo saltans KKT2 central domain | | 分子名称: | CHLORIDE ION, Protein kinase, putative, ... | | 著者 | Marciano, G, Nerusheva, O, Ishii, M, Akiyoshi, B. | | 登録日 | 2019-12-03 | | 公開日 | 2019-12-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Kinetoplastid kinetochore proteins KKT2 and KKT3 have unique centromere localization domains.
J.Cell Biol., 220, 2021
|
|
7NP4
 
 | | cAMP-bound rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | 著者 | Giese, H, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | 登録日 | 2021-02-26 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7NP3
 
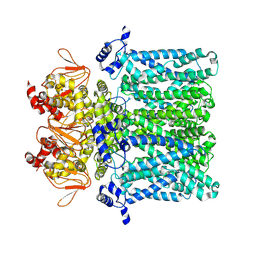 | | cAMP-free rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | 分子名称: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | 著者 | Giese, H.M, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | 登録日 | 2021-02-26 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7NH3
 
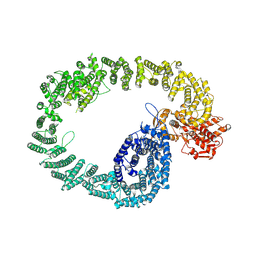 | | Nematocida Huwe1 in open conformation. | | 分子名称: | E3 ubiquitin-protein ligase HUWE1 | | 著者 | Petrova, O, Grishkovskaya, I, Grabarczyk, D.B, Kessler, D, Haselbach, D, Clausen, T. | | 登録日 | 2021-02-09 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (6.37 Å) | | 主引用文献 | Crystal structure of HUWE1: One ring to ubiquitinate them all
To Be Published
|
|
3LNE
 
 | |
3LNF
 
 | |
3LM0
 
 | | Crystal Structure of human Serine/Threonine Kinase 17B (STK17B) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Serine/threonine-protein kinase 17B, ... | | 著者 | Ugochukwu, E, Soundararajan, M, Rellos, P, Fedorov, O, Phillips, C, Wang, J, Hapka, E, Filippakopoulos, P, Chaikuad, A, Pike, A.C.W, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2010-01-29 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 |
|
|
3LNI
 
 | |
5VT1
 
 | | Crystal Structure of the Human CAMKK2B bound to a thiadiazinone benzamide inhibitor | | 分子名称: | 4-({5-[(3-hydroxy-4-methylphenyl)amino]-4-oxo-4H-1,2,6-thiadiazin-3-yl}amino)benzamide, Calcium/calmodulin-dependent protein kinase kinase 2, MAGNESIUM ION | | 著者 | Counago, R.M, Asquith, C.R.M, Arruda, P, Edwards, A.M, Gileadi, O, Kalogirou, A.S, Koutentis, P.A, Structural Genomics Consortium (SGC) | | 登録日 | 2017-05-15 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | 1,2,6-Thiadiazinones as Novel Narrow Spectrum Calcium/Calmodulin-Dependent Protein Kinase Kinase 2 (CaMKK2) Inhibitors.
Molecules, 23, 2018
|
|
3LNH
 
 | |
