5O1Q
 
 | | LysF1 sh3b domain structure | | Descriptor: | sh3b domain | | Authors: | Benesik, M, Novacek, J, Janda, L, Dopitova, R, Pernisova, M, Melkova, K, Tisakova, L, Doskar, J, Zidek, L, Hejatko, J, Pantucek, R. | | Deposit date: | 2017-05-19 | | Release date: | 2017-09-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Role of SH3b binding domain in a natural deletion mutant of Kayvirus endolysin LysF1 with a broad range of lytic activity.
Virus Genes, 54, 2018
|
|
7Z47
 
 | | Tail of bacteriophage SU10 | | Descriptor: | Adaptor protein, Putative structural protein, Putative tail fiber, ... | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-10 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
7Z4F
 
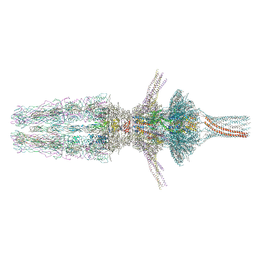 | | Tail of phage SU10 genome release intermediate | | Descriptor: | Adaptor protein, Portal protein, Putative structural protein, ... | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-17 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
7Z4A
 
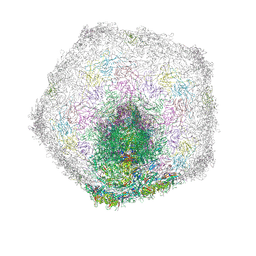 | | Bacteriophage SU10 tail and bottom part of the capsid (C1) | | Descriptor: | Adaptor protein, Major head protein, Portal protein, ... | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-17 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
7Z48
 
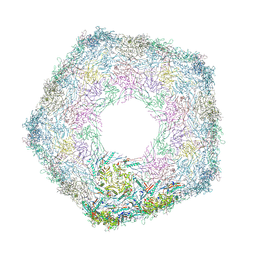 | | Bottom part (C5) of bacteriophage SU10 capsid | | Descriptor: | Major head protein | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-10 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
7Z44
 
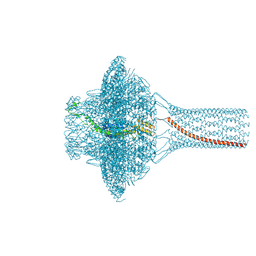 | | Portal of bacteriophage SU10 | | Descriptor: | Portal protein | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-10 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
7Z46
 
 | | Top part (C5) of bacteriophage SU10 capsid | | Descriptor: | Major head protein | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-24 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
7Z45
 
 | | Central part (C10) of bacteriophage SU10 capsid | | Descriptor: | Major head protein | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-24 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
7Z49
 
 | | The capsid of bacteriophage SU10. | | Descriptor: | Major head protein | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-10-05 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
7Z4B
 
 | | Bacteriophage SU10 virion (C1) | | Descriptor: | Adaptor, Major head protein, Portal protein, ... | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
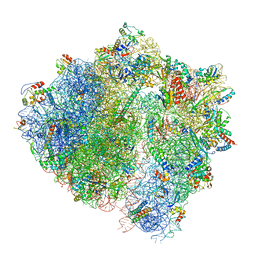
8CAI
 
 | | Streptomycin and Hygromycin B bound to the 30S body | | Descriptor: | 16S rRNA, 30S ribosomal protein S16, 30S ribosomal protein S2, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Novacek, J, Wilson, D.N. | | Deposit date: | 2023-01-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CGI
 
 | | Pentacycline TP038 bound to the 30S head | | Descriptor: | 16S rRNA, 30S ribosomal protein S7, Large ribosomal subunit protein bL31A, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Novacek, J, Wilson, D.N. | | Deposit date: | 2023-02-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (1.89 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CGU
 
 | | Gentamicin bound to the 30S body | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 16S rRNA, 23S rRNA, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Novacek, J, Wilson, D.N. | | Deposit date: | 2023-02-06 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (1.89 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CAM
 
 | | Evernimicin bound to the 50S subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L15, 50S ribosomal protein L25, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Novacek, J, Wilson, D.N. | | Deposit date: | 2023-01-24 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.86 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CGV
 
 | | Tiamulin bound to the 50S subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L15, 50S ribosomal protein L25, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Novacek, J, Wilson, D.N. | | Deposit date: | 2023-02-06 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.66 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6H05
 
 | | Cryo-electron microscopic structure of the dihydrolipoamide succinyltransferase (E2) component of the human alpha-ketoglutarate (2-oxoglutarate) dehydrogenase complex [residues 218-453] | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | | Authors: | Nagy, B, Zambo, Z, Hubert, A, Polak, M, Nemeria, N.S, Novacek, J, Jordan, F, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-07-06 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the dihydrolipoamide succinyltransferase (E2) component of the human alpha-ketoglutarate dehydrogenase complex (hKGDHc) revealed by cryo-EM and cross-linking mass spectrometry: Implications for the overall hKGDHc structure.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6HXX
 
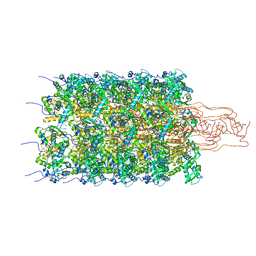 | | Potato virus Y | | Descriptor: | Coat protein, RNA (5'-R(P*UP*UP*UP*UP*U)-3') | | Authors: | Podobnik, M, Kezar, A, Novacek, J, Polak, M. | | Deposit date: | 2018-10-18 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the multitasking nature of the potato virus Y coat protein.
Sci Adv, 5, 2019
|
|
6HA8
 
 | | Cryo-EM structure of the ABCF protein VmlR bound to the Bacillus subtilis ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Graf, M, Huter, P, Abdelshahid, M, Novacek, J, Wilson, D.N. | | Deposit date: | 2018-08-07 | | Release date: | 2018-08-29 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for antibiotic resistance mediated by theBacillus subtilisABCF ATPase VmlR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8OOH
 
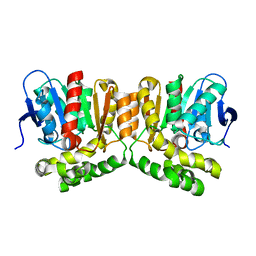 | |
5L8Q
 
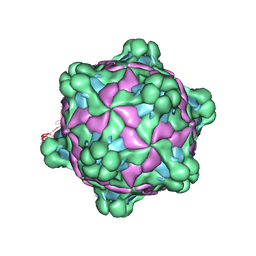 | | Structure of deformed wing virus, a honeybee pathogen | | Descriptor: | URIDINE-5'-MONOPHOSPHATE, VP1, VP2, ... | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2016-06-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5L7Q
 
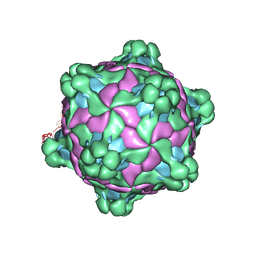 | | Structure of deformed wing virus, a honeybee pathogen | | Descriptor: | VP1, vp2, vp3 | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6HXZ
 
 | |
6HA1
 
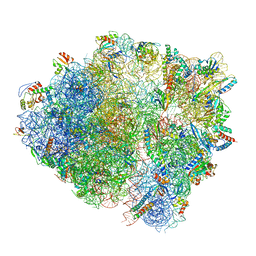 | | Cryo-EM structure of a 70S Bacillus subtilis ribosome translating the ErmD leader peptide in complex with telithromycin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Graf, M, Huter, P, Abdelshahid, M, Novacek, J, Wilson, D.N. | | Deposit date: | 2018-08-07 | | Release date: | 2018-08-29 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for antibiotic resistance mediated by theBacillus subtilisABCF ATPase VmlR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7BC3
 
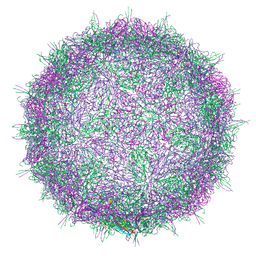 | | Native virion of Kashmir bee virus at acidic pH | | Descriptor: | Structural polyprotein | | Authors: | Mukhamedova, L, Plevka, P, Fuzik, T, Hrebik, D, Novacek, J. | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Virion structure and in vitro genome release mechanism of dicistrovirus Kashmir bee virus.
J.Virol., 95, 2021
|
|
7BGK
 
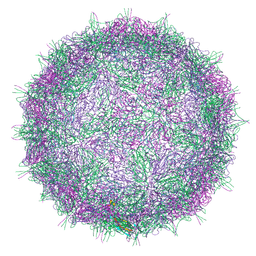 | | Native virion of Kashmir bee virus at neutral pH | | Descriptor: | Structural polyprotein | | Authors: | Mukhamedova, L, Plevka, P, Fuzik, T, Hrebik, D, Novacek, J. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Virion structure and in vitro genome release mechanism of dicistrovirus Kashmir bee virus.
J.Virol., 95, 2021
|
|
