1IV6
 
 | | Solution Structure of the DNA Complex of Human TRF1 | | Descriptor: | 5'-D(*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*C)-3', 5'-D(*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3', TELOMERIC REPEAT BINDING FACTOR 1 | | Authors: | Nishikawa, T, Okamura, H, Nagadoi, A, Konig, P, Rhodes, D, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-14 | | Release date: | 2002-04-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a telomeric DNA complex of human TRF1.
Structure, 9, 2001
|
|
1ITY
 
 | | Solution structure of the DNA binding domain of human TRF1 | | Descriptor: | TRF1 | | Authors: | Nishikawa, T, Okamura, H, Nagadoi, A, Konig, P, Rhodes, D, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-02-15 | | Release date: | 2002-03-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a telomeric DNA complex of human TRF1
Structure, 9, 2001
|
|
1X0S
 
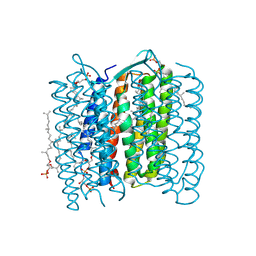 | | Crystal structure of the 13-cis isomer of bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nishikawa, T, Murakami, M, Kouyama, T. | | Deposit date: | 2005-03-28 | | Release date: | 2005-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the 13-cis isomer of bacteriorhodopsin in the dark-adapted state.
J.Mol.Biol., 352, 2005
|
|
1BA5
 
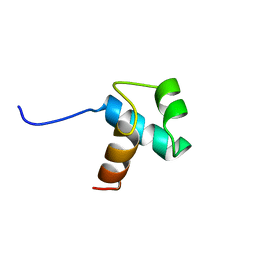 | | DNA-BINDING DOMAIN OF HUMAN TELOMERIC PROTEIN, HTRF1, NMR, 18 STRUCTURES | | Descriptor: | HTRF1 | | Authors: | Nishikawa, T, Nagadoi, A, Yoshimura, S, Aimoto, S, Nishimura, Y. | | Deposit date: | 1998-04-22 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of human telomeric protein, hTRF1.
Structure, 6, 1998
|
|
6PW7
 
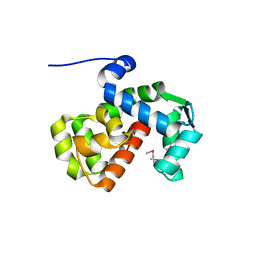 | | X-ray crystal structure of C. elegans STIM EF-SAM domain | | Descriptor: | CALCIUM ION, Stromal interaction molecule 1 | | Authors: | Enomoto, M, Nishikawa, T, Back, S.I, Ishiyama, N, Zheng, L, Stathopulos, P.B, Ikura, M. | | Deposit date: | 2019-07-22 | | Release date: | 2019-11-13 | | Last modified: | 2020-02-12 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Coordination of a Single Calcium Ion in the EF-hand Maintains the Off State of the Stromal Interaction Molecule Luminal Domain.
J.Mol.Biol., 432, 2020
|
|
6CC9
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-06 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
1UCQ
 
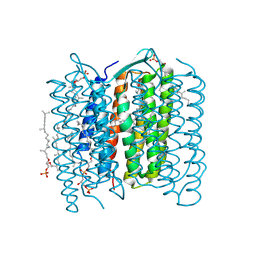 | | Crystal structure of the L intermediate of bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, RETINAL, ... | | Authors: | Kouyama, T, Nishikawa, T, Tokuhisa, T, Okumura, H. | | Deposit date: | 2003-04-17 | | Release date: | 2003-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the L Intermediate of Bacteriorhodopsin: Evidence for Vertical Translocation of a Water Molecule during the Proton Pumping Cycle.
J.Mol.Biol., 335, 2004
|
|
6CCX
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6CCH
 
 | | NMR data-driven model of GTPase KRas-GMPPNP tethered to a nanodisc (E3 state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
