2ZOE
 
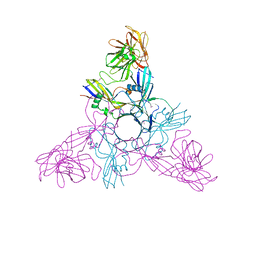 | | HA3 subcomponent of Clostridium botulinum type C progenitor toxin, complex with N-acetylneuramic acid | | Descriptor: | Hemagglutinin components HA3, N-acetyl-beta-neuraminic acid | | Authors: | Nakamura, T, Kotani, M, Tonozuka, T, Ide, A, Oguma, K, Nishikawa, A. | | Deposit date: | 2008-05-09 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the HA3 Subcomponent of Clostridium botulinum Type C Progenitor Toxin
J.Mol.Biol., 385, 2009
|
|
2ZYK
 
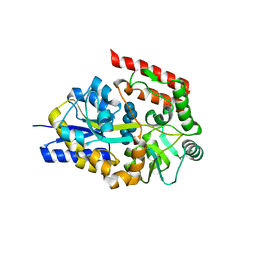 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with gamma-cyclodextrin | | Descriptor: | Cyclooctakis-(1-4)-(alpha-D-glucopyranose), Solute-binding protein | | Authors: | Tonozuka, T, Sogawa, A, Yamada, M, Matsumoto, N, Yoshida, H, Kamitori, S, Ichikawa, K, Mizuno, M, Nishikawa, A, Sakano, Y. | | Deposit date: | 2009-01-26 | | Release date: | 2009-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for cyclodextrin recognition by Thermoactinomyces vulgaris cyclo/maltodextrin-binding protein
Febs J., 274, 2007
|
|
1ULV
 
 | | Crystal Structure of Glucodextranase Complexed with Acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, glucodextranase | | Authors: | Mizuno, M, Tonozuka, T, Suzuki, S, Uotsu-Tomita, R, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2003-09-16 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural insights into substrate specificity and function of glucodextranase
J.Biol.Chem., 279, 2004
|
|
3AJ5
 
 | | HA1 (HA33) subcomponent of botulinum type C progenitor toxin complexed with N-acetylgalactosamine, bound at site II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Main hemagglutinin component | | Authors: | Nakamura, T, Tonozuka, T, Sato, R, Oguma, K, Nishikawa, A. | | Deposit date: | 2010-05-24 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular diversity of the two sugar-binding sites of the beta-trefoil lectin HA33/C (HA1) from Clostridium botulinum type C neurotoxin
Arch.Biochem.Biophys., 512, 2011
|
|
3AJ6
 
 | | HA1 (HA33) mutant F179I of botulinum type C progenitor toxin complexed with N-acetylgalactosamine, bound at site II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Main hemagglutinin component | | Authors: | Nakamura, T, Tonozuka, T, Sato, R, Oguma, K, Nishikawa, A. | | Deposit date: | 2010-05-24 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Molecular diversity of the two sugar-binding sites of the beta-trefoil lectin HA33/C (HA1) from Clostridium botulinum type C neurotoxin
Arch.Biochem.Biophys., 512, 2011
|
|
3A64
 
 | | Crystal structure of CcCel6C, a glycoside hydrolase family 6 enzyme, from Coprinopsis cinerea | | Descriptor: | Cellobiohydrolase, MAGNESIUM ION | | Authors: | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
3A9B
 
 | | CcCel6C, a glycoside hydrolase family 6 enzyme, complexed with cellobiose | | Descriptor: | Cellobiohydrolase, MAGNESIUM ION, beta-D-glucopyranose, ... | | Authors: | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
3ABX
 
 | | CcCel6C, a glycoside hydrolase family 6 enzyme, complexed with p-nitrophenyl beta-D-cellotrioside | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranoside, Cellobiohydrolase, MAGNESIUM ION | | Authors: | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-12-24 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
3VSR
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase catalytic domain | | Descriptor: | Beta-fructofuranosidase | | Authors: | Tonozuka, T, Tamaki, A, Yokoi, G, Miyazaki, T, Ichikawa, M, Nishikawa, A, Ohta, Y, Hidaka, Y, Katayama, K, Hatada, Y, Ito, T, Fujita, K. | | Deposit date: | 2012-05-08 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a lactosucrose-producing enzyme, Arthrobacter sp. K-1 beta-fructofuranosidase
Enzyme.Microb.Technol., 51, 2012
|
|
3W7T
 
 | | Escherichia coli K12 YgjK complexed with mannose | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Uncharacterized protein YgjK, ... | | Authors: | Miyazaki, T, Kurakata, Y, Uechi, A, Yoshida, H, Kamitori, S, Sakano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2013-03-06 | | Release date: | 2013-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the substrate specificity and function of Escherichia coli K12 YgjK, a glucosidase belonging to the glycoside hydrolase family 63.
J.Mol.Biol., 381, 2008
|
|
3VSS
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase catalytic domain complexed with fructose | | Descriptor: | Beta-fructofuranosidase, beta-D-fructofuranose | | Authors: | Tonozuka, T, Tamaki, A, Yokoi, G, Miyazaki, T, Ichikawa, M, Nishikawa, A, Ohta, Y, Hidaka, Y, Katayama, K, Hatada, Y, Ito, T, Fujita, K. | | Deposit date: | 2012-05-08 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of a lactosucrose-producing enzyme, Arthrobacter sp. K-1 beta-fructofuranosidase
Enzyme.Microb.Technol., 51, 2012
|
|
3W7S
 
 | | Escherichia coli K12 YgjK complexed with glucose | | Descriptor: | CALCIUM ION, Uncharacterized protein YgjK, alpha-D-glucopyranose | | Authors: | Miyazaki, T, Kurakata, Y, Uechi, A, Yoshida, H, Kamitori, S, Sakano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2013-03-06 | | Release date: | 2013-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the substrate specificity and function of Escherichia coli K12 YgjK, a glucosidase belonging to the glycoside hydrolase family 63.
J.Mol.Biol., 381, 2008
|
|
3W7U
 
 | | Escherichia coli K12 YgjK complexed with galactose | | Descriptor: | CALCIUM ION, Uncharacterized protein YgjK, alpha-D-galactopyranose | | Authors: | Miyazaki, T, Kurakata, Y, Uechi, A, Yoshida, H, Kamitori, S, Sakano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2013-03-06 | | Release date: | 2013-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into the substrate specificity and function of Escherichia coli K12 YgjK, a glucosidase belonging to the glycoside hydrolase family 63.
J.Mol.Biol., 381, 2008
|
|
3VOF
 
 | | Cellobiohydrolase mutant, CcCel6C D102A, in the closed form | | Descriptor: | Cellobiohydrolase, beta-D-glucopyranose | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-23 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3VOH
 
 | | CcCel6A catalytic domain complexed with cellobiose | | Descriptor: | Cellobiohydrolase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3VOJ
 
 | | CcCel6A catalytic domain mutant D164A | | Descriptor: | Cellobiohydrolase | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3VOG
 
 | | Catalytic domain of the cellobiohydrolase, CcCel6A, from Coprinopsis cinerea | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cellobiohydrolase | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3VOI
 
 | | CcCel6A catalytic domain complexed with p-nitrophenyl beta-D-cellotrioside | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranoside, Cellobiohydrolase, MAGNESIUM ION | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3W7W
 
 | | Crystal structure of E. coli YgjK E727A complexed with 2-O-alpha-D-glucopyranosyl-alpha-D-galactopyranose | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Uncharacterized protein YgjK, ... | | Authors: | Miyazaki, T, Ichikawa, M, Yokoi, G, Kitaoka, M, Mori, H, Kitano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2013-03-08 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a bacterial glycoside hydrolase family 63 enzyme in complex with its glycosynthase product, and insights into the substrate specificity.
Febs J., 280, 2013
|
|
3WPU
 
 | | Full-length beta-fructofuranosidase from Microbacterium saccharophilum K-1 | | Descriptor: | Beta-fructofuranosidase, GLYCEROL | | Authors: | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-01-17 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3W9A
 
 | | Crystal structure of the catalytic domain of the glycoside hydrolase family 131 protein from Coprinopsis cinerea | | Descriptor: | GLYCEROL, Putative uncharacterized protein | | Authors: | Miyazaki, T, Tanaka, Y, Tamura, M, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2013-04-01 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of the N-terminal domain of a glycoside hydrolase family 131 protein from Coprinopsis cinerea
Febs Lett., 587, 2013
|
|
3WPV
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase mutant T47S/F447V/F470Y/P500S | | Descriptor: | Beta-fructofuranosidase, GLYCEROL | | Authors: | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-01-17 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3WPZ
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase mutant T47S/S200T/F447P/F470Y/P500S | | Descriptor: | Beta-fructofuranosidase | | Authors: | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-01-17 | | Release date: | 2014-03-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3WZ1
 
 | | Catalytic domain of beta-agarase from Microbulbifer thermotolerans JAMB-A94 | | Descriptor: | Agarase, GLYCEROL, SODIUM ION | | Authors: | Takagi, E, Hatada, Y, Akita, M, Ohta, Y, Yokoi, G, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the catalytic domain of a GH16 beta-agarase from a deep-sea bacterium, Microbulbifer thermotolerans JAMB-A94
Biosci.Biotechnol.Biochem., 79, 2015
|
|
3WWG
 
 | | Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isopullulanase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Miyazaki, T, Yashiro, H, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-06-17 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The side chain of a glycosylated asparagine residue is important for the stability of isopullulanase
J.Biochem., 157, 2015
|
|
