1S74
 
 | | SOLUTION STRUCTURE OF A DNA DUPLEX CONTAINING AN ALPHA-ANOMERIC ADENOSINE: INSIGHTS INTO SUBSTRATE RECOGNITION BY ENDONUCLEASE IV | | Descriptor: | 5'-D(*CP*GP*TP*CP*GP*TP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*(A3A)P*CP*GP*AP*CP*G)-3' | | Authors: | Aramini, J.M, Cleaver, S.H, Pon, R.T, Cunningham, R.P, Germann, M.W. | | Deposit date: | 2004-01-28 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing an alpha-Anomeric Adenosine: Insights into Substrate Recognition by Endonuclease IV.
J.Mol.Biol., 338, 2004
|
|
7L91
 
 | | Structure of Metallo Beta-Lactamase L1 in a Complex with Hydrolyzed Moxalactam Determined by Pink-Beam Serial Crystallography | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Vukica, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-01 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7KOA
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Shuvalova, L, Lavens, A, Henning, R, Maltseva, N, Rosas-Lemus, M, Kim, Y, Satchell, K.J.F, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography
To Be Published
|
|
4J94
 
 | | Crystal structure of MycP1 from the ESX-1 type VII secretion system | | Descriptor: | Membrane-anchored mycosin mycp1 | | Authors: | Solomonson, M, Wasney, G.A, Watanabe, N, Gruninger, R.J, Prehna, G, Strynadka, N.C.J. | | Deposit date: | 2013-02-15 | | Release date: | 2013-05-01 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Structure of the Mycosin-1 Protease from the Mycobacterial ESX-1 Protein Type VII Secretion System.
J.Biol.Chem., 288, 2013
|
|
4KPG
 
 | | Crystal structure of MycP1 from the ESX-1 type VII secretion system | | Descriptor: | Membrane-anchored mycosin mycp1 | | Authors: | Solomonson, M, Wasney, G.A, Watanabe, N, Gruninger, R.J, Prehna, G, Strynadka, N.C.J. | | Deposit date: | 2013-05-13 | | Release date: | 2013-05-22 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Structure of the Mycosin-1 Protease from the Mycobacterial ESX-1 Protein Type VII Secretion System.
J.Biol.Chem., 288, 2013
|
|
1QUM
 
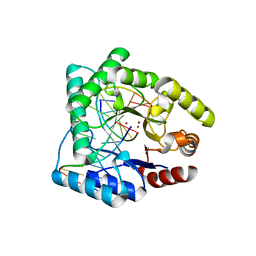 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI ENDONUCLEASE IV IN COMPLEX WITH DAMAGED DNA | | Descriptor: | 5'-D(*(3DR)P*CP*GP*AP*CP*GP*A)-3', 5'-D(*CP*GP*TP*CP*C)-3', 5'-D(*TP*CP*GP*TP*CP*GP*GP*GP*GP*AP*CP*G)-3', ... | | Authors: | Hosfield, D.J, Guan, Y, Haas, B.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1999-07-01 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis.
Cell(Cambridge,Mass.), 98, 1999
|
|
1QTW
 
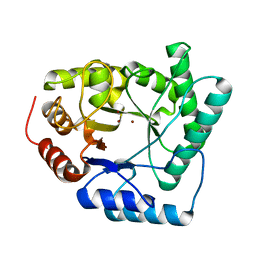 | | HIGH-RESOLUTION CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI DNA REPAIR ENZYME ENDONUCLEASE IV | | Descriptor: | ENDONUCLEASE IV, ZINC ION | | Authors: | Hosfield, D.J, Guan, Y, Haas, B.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1999-06-29 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis.
Cell(Cambridge,Mass.), 98, 1999
|
|
1KEA
 
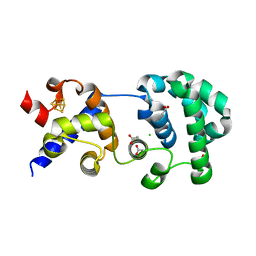 | | STRUCTURE OF A THERMOSTABLE THYMINE-DNA GLYCOSYLASE | | Descriptor: | ACETATE ION, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Mol, C.D, Arvai, A.S, Begley, T.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 2001-11-14 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of a thermostable thymine-DNA glycosylase: evidence for base twisting to remove mismatched normal DNA bases.
J.Mol.Biol., 315, 2002
|
|
3UMD
 
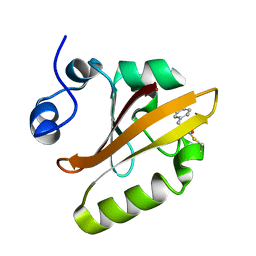 | | Structure of pB intermediate of Photoactive yellow protein (PYP) at pH 4. | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Tripathi, S, Srajer, V, Purwar, N, Henning, R, Schmidt, M. | | Deposit date: | 2011-11-13 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | pH Dependence of the Photoactive Yellow Protein Photocycle Investigated by Time-Resolved Crystallography.
Biophys.J., 102, 2012
|
|
5AJ2
 
 | | Cryo electron tomography of the Naip5-Nlrc4 inflammasome | | Descriptor: | NLR FAMILY CARD DOMAIN-CONTAINING PROTEIN 4 | | Authors: | Diebolder, C.A, Halff, E.F, Koster, A.J, Huizinga, E.G, Koning, R.I. | | Deposit date: | 2015-02-20 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (40 Å) | | Cite: | Cryoelectron Tomography of the Naip5/Nlrc4 Inflammasome: Implications for Nlr Activation.
Structure, 23, 2015
|
|
4LBV
 
 | | Identifying ligand binding hot spots in proteins using brominated fragments | | Descriptor: | AMMONIUM ION, CHLORIDE ION, Elongation factor Tu-A, ... | | Authors: | Groftehauge, M.K, Therkelsen, M, Taaning, R, Skrydstrup, T, Morth, J.P, Nissen, P. | | Deposit date: | 2013-06-21 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identifying ligand-binding hot spots in proteins using brominated fragments.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3UME
 
 | | Structure of pB intermediate of Photoactive yellow protein (PYP) at pH 7 | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Tripathi, S, Srajer, V, Purwar, N, Henning, R, Schmidt, M. | | Deposit date: | 2011-11-13 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | pH Dependence of the Photoactive Yellow Protein Photocycle Investigated by Time-Resolved Crystallography.
Biophys.J., 102, 2012
|
|
4LC0
 
 | | Identifying ligand binding hot spots in proteins using brominated fragments | | Descriptor: | AMMONIUM ION, Elongation factor Tu-A, MAGNESIUM ION, ... | | Authors: | Groftehauge, M.K, Therkelsen, M, Taaning, R, Skrydstrup, T, Morth, J.P, Nissen, P. | | Deposit date: | 2013-06-21 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.221 Å) | | Cite: | Identifying ligand-binding hot spots in proteins using brominated fragments.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4LBW
 
 | | Identifying ligand binding hot spots in proteins using brominated fragments | | Descriptor: | AMMONIUM ION, Elongation factor Tu-A, MAGNESIUM ION, ... | | Authors: | Groftehauge, M.K, Therkelsen, M, Taaning, R, Skrydstrup, T, Morth, J.P, Nissen, P. | | Deposit date: | 2013-06-21 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.741 Å) | | Cite: | Identifying ligand-binding hot spots in proteins using brominated fragments.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4LBZ
 
 | | Identifying ligand binding hot spots in proteins using brominated fragments | | Descriptor: | AMMONIUM ION, Elongation factor Tu-A, MAGNESIUM ION, ... | | Authors: | Groftehauge, M.K, Therkelsen, M, Taaning, R, Skrydstrup, T, Morth, J.P, Nissen, P. | | Deposit date: | 2013-06-21 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.223 Å) | | Cite: | Identifying ligand-binding hot spots in proteins using brominated fragments.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4LBY
 
 | | Identifying ligand binding hot spots in proteins using brominated fragments | | Descriptor: | AMMONIUM ION, Elongation factor Tu-A, MAGNESIUM ION, ... | | Authors: | Groftehauge, M.K, Therkelsen, M, Taaning, R, Skrydstrup, T, Morth, J.P, Nissen, P. | | Deposit date: | 2013-06-21 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Identifying ligand-binding hot spots in proteins using brominated fragments.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6FCZ
 
 | | Model of gC1q-Fc complex based on 7A EM map | | Descriptor: | Complement C1q subcomponent subunit A, Complement C1q subcomponent subunit B, Complement C1q subcomponent subunit C, ... | | Authors: | Ugurlar, D, Howes, S.C, de Kreuk, B.J.K, de Jong, R.N, Beurskens, F.J, Koster, A.J, Parren, P.W.H.I, Sharp, T.H, Gros, P, Koning, R.I. | | Deposit date: | 2017-12-21 | | Release date: | 2018-02-28 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structures of C1-IgG1 provide insights into how danger pattern recognition activates complement.
Science, 359, 2018
|
|
2IY3
 
 | | Structure of the E. Coli Signal Regognition Particle | | Descriptor: | 4.5S RNA, SIGNAL SEQUENCE, Signal recognition particle protein,Signal recognition particle 54 kDa protein | | Authors: | Schaffitzel, C, Oswald, M, Berger, I, Ishikawa, T, Abrahams, J.P, Koerten, H.K, Koning, R.I, Ban, N. | | Deposit date: | 2006-07-12 | | Release date: | 2006-11-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structure of the E. Coli Signal Recognition Particle Bound to a Translating Ribosome
Nature, 444, 2006
|
|
7Z18
 
 | | E. coli C-P lyase bound to a PhnK ABC dimer and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7Z19
 
 | | E. coli C-P lyase bound to a single PhnK ABC domain | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7Z15
 
 | | E. coli C-P lyase bound to a PhnK/PhnL dual ABC dimer and ADP + Pi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7Z16
 
 | | E. coli C-P lyase bound to PhnK/PhnL dual ABC dimer with AMPPNP and PhnK E171Q mutation | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnI, ... | | Authors: | Amstrup, S.K, Sofus, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7Z17
 
 | | E. coli C-P lyase bound to a PhnK ABC dimer in an open conformation | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
2LIB
 
 | | DNA sequence context conceals alpha anomeric lesion | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*TP*GP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*(A3A)P*GP*GP*AP*CP*G)-3') | | Authors: | Johnson, C.N, Spring, A.M, Cunningham, R.P, Germann, M.W. | | Deposit date: | 2011-08-27 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | DNA sequence context conceals alpha-anomeric lesions.
J.Mol.Biol., 416, 2012
|
|
