8IZU
 
 | |
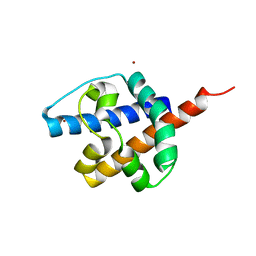
8IZT
 
 | |
7WZO
 
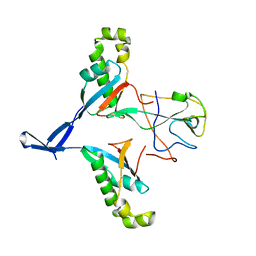 | |
9FGO
 
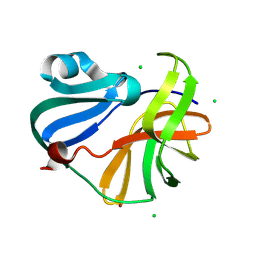 | | Crystal structure of Enterovirus 71 2A protease mutant C110A containing VP1-2A junction in the active site | | Descriptor: | CHLORIDE ION, Polyprotein, ZINC ION | | Authors: | Ni, X, Koekemoer, L, Williams, E.P, Wang, S, Wright, N.D, Godoy, A.S, Aschenbrenner, J.C, Balcomb, B.H, Lithgo, R.M, Marples, P.G, Fairhead, M, Thompson, W, Kirkegaard, K, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-05-24 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of Enterovirus 71 2A protease mutant C110A containing VP1-2A junction in the active site
To Be Published
|
|
7Q8A
 
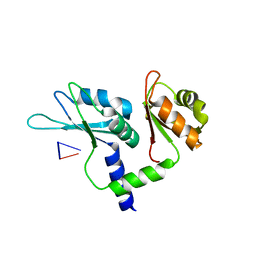 | | Crystal structure of tandem domain RRM1-2 of FUBP-interacting repressor (FIR) bound to FUSE ssDNA fragment | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*GP*T)-3'), Poly(U)-binding-splicing factor PUF60, ... | | Authors: | Ni, X, Joerger, A.C, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-10 | | Release date: | 2022-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of tandem domain RRM1-2 of FIR bound to FUSE ssDNA fragment
To Be Published
|
|
6OSL
 
 | |
7Z3X
 
 | | Crystal structure of FIR RRM1-2 Y115F mutant bound to FUSE ssDNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*T)-3'), DNA (5'-D(P*GP*TP*())-3'), ... | | Authors: | Ni, X, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-02 | | Release date: | 2022-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of FIR RRM1-2 Y115F mutant bound to FUSE ssDNA
To Be Published
|
|
8C1L
 
 | | Crystal structure of HNF4 alpha LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | 1,2-ETHANEDIOL, Hepatocyte nuclear factor 4-alpha, Nuclear receptor coactivator 2, ... | | Authors: | Ni, X, Merk, D, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of HNF4 alpha LBD in complexes with palmitic acid and GRIP-1 peptide
To Be Published
|
|
6WUJ
 
 | | Mitochondrial SAM complex - monomer in detergent | | Descriptor: | Bac_surface_Ag domain-containing protein, Sam35, Tom37 domain-containing protein | | Authors: | Ni, X, Botos, I, Diederichs, K. | | Deposit date: | 2020-05-04 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insight into mitochondrial beta-barrel outer membrane protein biogenesis.
Nat Commun, 11, 2020
|
|
6WUT
 
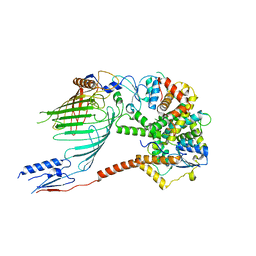 | |
6WUH
 
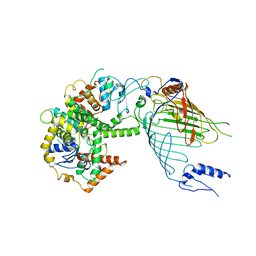 | | Mitochondrial SAM complex in lipid nanodiscs | | Descriptor: | Bac_surface_Ag domain-containing protein, Sam35, Tom37 domain-containing protein | | Authors: | Ni, X, Botos, I, Diederichs, K. | | Deposit date: | 2020-05-04 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insight into mitochondrial beta-barrel outer membrane protein biogenesis.
Nat Commun, 11, 2020
|
|
6WUM
 
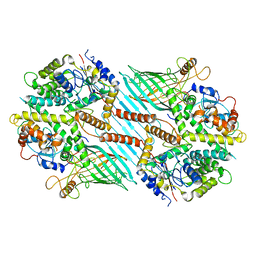 | | Mitochondrial SAM complex - dimer 2 in detergent | | Descriptor: | Bac_surface_Ag domain-containing protein, Sam35, Tom37 domain-containing protein | | Authors: | Ni, X, Botos, I, Diederichs, K. | | Deposit date: | 2020-05-04 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into mitochondrial beta-barrel outer membrane protein biogenesis.
Nat Commun, 11, 2020
|
|
6WUN
 
 | |
7P4E
 
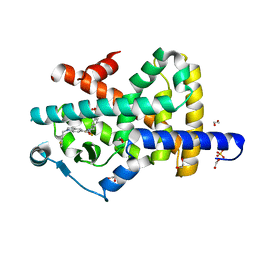 | | Crystal structure of PPARgamma in complex with compound FL217 | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma, SULFATE ION, ... | | Authors: | Ni, X, Lillich, F, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of Dual Partial Peroxisome Proliferator-Activated Receptor gamma Agonists/Soluble Epoxide Hydrolase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7P4K
 
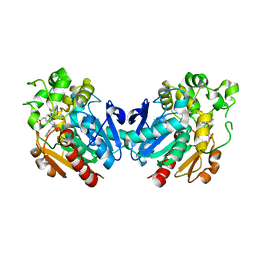 | | Soluble epoxide hydrolase in complex with FL217 | | Descriptor: | Bifunctional epoxide hydrolase 2, ~{N}-[[4-(cyclopropylsulfonylamino)-2-(trifluoromethyl)phenyl]methyl]-1-[(3-fluorophenyl)methyl]indole-5-carboxamide | | Authors: | Ni, X, Kramer, J.S, Lillich, F, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of Dual Partial Peroxisome Proliferator-Activated Receptor gamma Agonists/Soluble Epoxide Hydrolase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7A7G
 
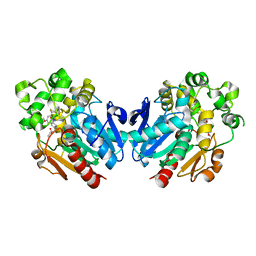 | | Soluble epoxide hydrolase in complex with TK90 | | Descriptor: | (2~{R})-2-[[4-[[4-methoxy-2-(trifluoromethyl)phenyl]methylcarbamoyl]phenyl]methyl]butanoic acid, Bifunctional epoxide hydrolase 2 | | Authors: | Ni, X, Kramer, J.S, Kirchner, T, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Combined Cardioprotective and Adipocyte Browning Effects Promoted by the Eutomer of Dual sEH/PPAR gamma Modulator.
J.Med.Chem., 64, 2021
|
|
7A7H
 
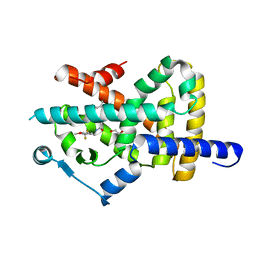 | | Crystal structure of PPARgamma in complex with compound TK90 | | Descriptor: | (2~{R})-2-[[4-[[4-methoxy-2-(trifluoromethyl)phenyl]methylcarbamoyl]phenyl]methyl]butanoic acid, 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma | | Authors: | Ni, X, Kirchner, T, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Combined Cardioprotective and Adipocyte Browning Effects Promoted by the Eutomer of Dual sEH/PPAR gamma Modulator.
J.Med.Chem., 64, 2021
|
|
7B0T
 
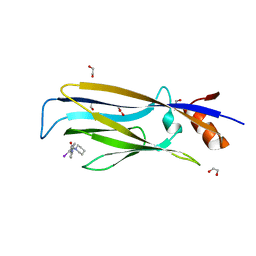 | | Crystal structure of MLLT1 YEATS domain T3 mutant in complex with benzimidazole-amide based compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 3-iodanyl-4-methyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Ni, X, Chaikuad, A, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-21 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and Inhibitor Binding Characterization of Oncogenic MLLT1 Mutants.
Acs Chem.Biol., 16, 2021
|
|
6Y2C
 
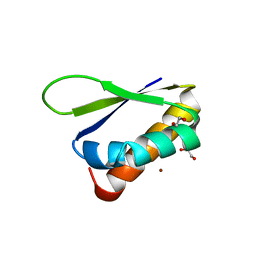 | | Crystal structure of the third KH domain of FUBP1 | | Descriptor: | 1,2-ETHANEDIOL, Far upstream element-binding protein 1, ZINC ION | | Authors: | Ni, X, Joerger, A.C, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
6Y24
 
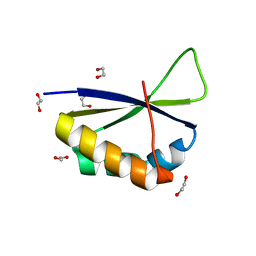 | | Crystal structure of fourth KH domain of FUBP1 | | Descriptor: | 1,2-ETHANEDIOL, Far upstream element-binding protein 1 | | Authors: | Ni, X, Joerger, A.C, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
7BF3
 
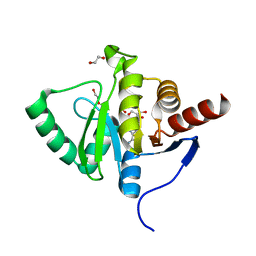 | | Crystal structure of SARS-CoV-2 macrodomain in complex with adenosine | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, MAGNESIUM ION, ... | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
7BF6
 
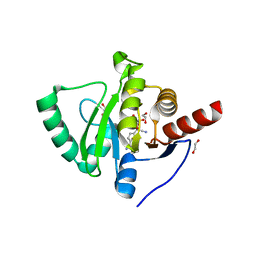 | | Crystal structure of SARS-CoV-2 macrodomain in complex with remdesivir metabolite GS-441524 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile, 1,2-ETHANEDIOL, Papain-like protease nsp3 | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
7BF4
 
 | | Crystal structure of SARS-CoV-2 macrodomain in complex with GMP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-MONOPHOSPHATE, NSP3 macrodomain | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6WUL
 
 | | Mitochondrial SAM complex - dimer 1 in detergent | | Descriptor: | Bac_surface_Ag domain-containing protein, Sam35, Tom37 domain-containing protein | | Authors: | Ni, X, Botos, I, Diederichs, K. | | Deposit date: | 2020-05-04 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into mitochondrial beta-barrel outer membrane protein biogenesis.
Nat Commun, 11, 2020
|
|
6Y2D
 
 | | Crystal structure of the second KH domain of FUBP1 | | Descriptor: | Far upstream element-binding protein 1, GLYCEROL, SULFATE ION | | Authors: | Ni, X, Chaikuad, A, Joerger, A.C, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
