2O67
 
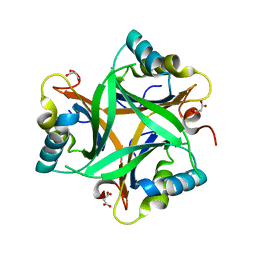 | | Crystal structure of Arabidopsis thaliana PII bound to malonate | | Descriptor: | MALONATE ION, PII protein | | Authors: | Mizuno, Y.M, Berenger, B, Moorhead, G.B.G, Ng, K.K.S. | | Deposit date: | 2006-12-06 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Arabidopsis PII Reveals Novel Structural Elements Unique to Plants.
Biochemistry, 46, 2007
|
|
2O66
 
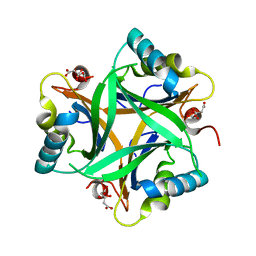 | | Crystal Structure of Arabidopsis thaliana PII bound to citrate | | Descriptor: | CITRATE ANION, PII protein | | Authors: | Mizuno, Y, Berenger, B, Moorhead, G.B.G, Ng, K.K.S. | | Deposit date: | 2006-12-06 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Arabidopsis PII Reveals Novel Structural Elements Unique to Plants.
Biochemistry, 46, 2007
|
|
4FIY
 
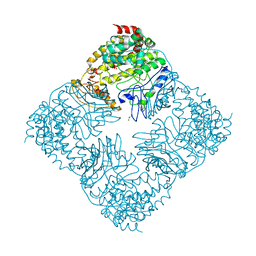 | | Crystal Structure of GlfT2 Complexed with UDP | | Descriptor: | GLYCEROL, MANGANESE (II) ION, UDP-galactofuranosyl transferase GlfT2, ... | | Authors: | Wheatley, R.W, Zheng, R.B, Lowary, T.L, Ng, K.K.S. | | Deposit date: | 2012-06-11 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Tetrameric Structure of the GlfT2 Galactofuranosyltransferase Reveals a Scaffold for the Assembly of Mycobacterial Arabinogalactan.
J.Biol.Chem., 287, 2012
|
|
4FIX
 
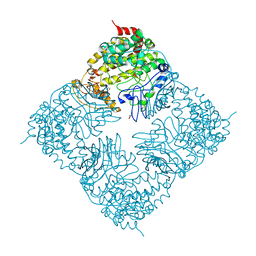 | | Crystal Structure of GlfT2 | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Wheatley, R.W, Zheng, R.B, Lowary, T.L, Ng, K.K.S. | | Deposit date: | 2012-06-11 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Tetrameric Structure of the GlfT2 Galactofuranosyltransferase Reveals a Scaffold for the Assembly of Mycobacterial Arabinogalactan.
J.Biol.Chem., 287, 2012
|
|
1G3C
 
 | | BOVINE BETA-TRYPSIN BOUND TO PARA-AMIDINO SCHIFF BASE IRON(III) CHELATE | | Descriptor: | 2-(4-CARBAMIMIDOYL-2-HYDROXY-BENZYLAMINO)-PROPIONIC ACID, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Toyota, E, Ng, K.K.S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of complexes between bovine beta-trypsin and Schiff base copper(II) or iron(III) chelates.
J.Mol.Biol., 305, 2001
|
|
1G3E
 
 | | BOVINE BETA-TRYPSIN BOUND TO PARA-AMIDINO SCHIFF-BASE COPPER (II) CHELATE | | Descriptor: | 2-(4-CARBAMIMIDOYL-2-HYDROXY-BENZYLAMINO)-PROPIONIC ACID, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Toyota, E, Ng, K.K.S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of complexes between bovine beta-trypsin and Schiff base copper(II) or iron(III) chelates.
J.Mol.Biol., 305, 2001
|
|
1G3D
 
 | | BOVINE BETA-TRYPSIN BOUND TO META-AMIDINO SCHIFF BASE COPPER (II) CHELATE | | Descriptor: | 2-(5-CARBAMIMIDOYL-2-HYDROXY-BENZYLAMINO)-PROPIONIC ACID, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Toyota, E, Ng, K.K.S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of complexes between bovine beta-trypsin and Schiff base copper(II) or iron(III) chelates.
J.Mol.Biol., 305, 2001
|
|
1G3B
 
 | | BOVINE BETA-TRYPSIN BOUND TO META-AMIDINO SCHIFF BASE MAGNESIUM(II) CHELATE | | Descriptor: | 2-(5-CARBAMIMIDOYL-2-HYDROXY-BENZYLAMINO)-PROPIONIC ACID, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Toyota, E, Ng, K.K.S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of complexes between bovine beta-trypsin and Schiff base copper(II) or iron(III) chelates.
J.Mol.Biol., 305, 2001
|
|
3HNV
 
 | | CS-35 Fab Complex with Oligoarabinofuranosyl Tetrasaccharide (branch part of Hexasaccharide) | | Descriptor: | CS-35 Fab Heavy Chain, CS-35 Fab Light Chain, beta-D-arabinofuranose-(1-2)-alpha-D-arabinofuranose-(1-3)-alpha-D-arabinofuranose-(1-5)-methyl alpha-D-arabinofuranoside | | Authors: | Murase, T, Zheng, R.B, Joe, M, Bai, Y, Marcus, S.L, Lowary, T.L, Ng, K.K.S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into antibody recognition of mycobacterial polysaccharides.
J.Mol.Biol., 392, 2009
|
|
3HNT
 
 | | CS-35 Fab complex with a linear, terminal oligoarabinofuranosyl tetrasaccharide from lipoarabinomannan | | Descriptor: | CS-35 Fab Heavy Chain, CS-35 Fab Light Chain, beta-D-arabinofuranose-(1-2)-alpha-D-arabinofuranose-(1-5)-alpha-D-arabinofuranose-(1-5)-methyl alpha-D-arabinofuranoside | | Authors: | Murase, T, Zheng, R.B, Joe, M, Bai, Y, Marcus, S.L, Lowary, T.L, Ng, K.K.S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into antibody recognition of mycobacterial polysaccharides.
J.Mol.Biol., 392, 2009
|
|
4NBY
 
 | | Crystal Structure of TcdA-A2 Bound to Two Molecules of A20.1 VHH | | Descriptor: | A20.1 VHH, Cell wall-binding repeat protein | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
4NC1
 
 | | Crystal Structure of TcdA-A2 Bound to A20.1 VHH and A26.8 VHH | | Descriptor: | A20.1 VHH, A26.8 VHH, Cell wall-binding repeat protein | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
5KOC
 
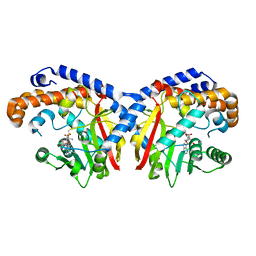 | | Pavine N-methyltransferase in complex with S-adenosylmethionine pH 7 | | Descriptor: | Pavine N-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Torres, M.A, Hoffarth, E, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
5KOK
 
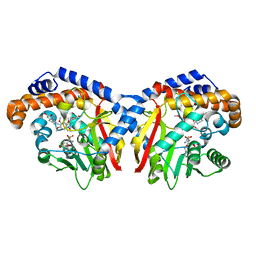 | | Pavine N-methyltransferase in complex with Tetrahydropapaverine and S-adenosylhomocysteine pH 7.25 | | Descriptor: | (1~{R})-1-[(3,4-dimethoxyphenyl)methyl]-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline, (1~{S})-1-[(3,4-dimethoxyphenyl)methyl]-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline, Pavine N-methyltransferase, ... | | Authors: | Torres, M.A, Hoffarth, E, Eugenio, L, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
4NC2
 
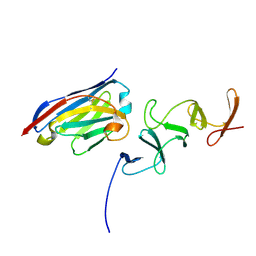 | | Crystal structure of TcdB-B1 bound to B39 VHH | | Descriptor: | B39 VHH, Toxin B | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
5KN4
 
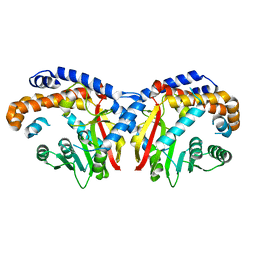 | | Pavine N-methyltransferase apoenzyme pH 6.0 | | Descriptor: | Pavine N-methyltransferase | | Authors: | Torres, M.A, Hoffarth, E, Eugenio, L, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-06-27 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
5KPC
 
 | | Pavine N-methyltransferase H206A mutant in complex with S-adenosylmethionine pH 6 | | Descriptor: | Pavine N-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Torres, M.A, Hoffarth, E, Eugenio, L, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-07-03 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
5KPG
 
 | | Pavine N-methyltransferase in complex with S-adenosylhomocysteine pH 7 | | Descriptor: | Pavine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Torres, M.A, Hoffarth, E, Eugenio, L, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-07-04 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
4NBX
 
 | | Crystal Structure of Clostridium difficile Toxin A fragment TcdA-A1 Bound to A20.1 VHH | | Descriptor: | A20.1 VHH, TcdA | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
3HNS
 
 | | CS-35 Fab Complex with Oligoarabinofuranosyl Hexasaccharide | | Descriptor: | CS-35 Fab Heavy Chain, CS-35 Fab Light Chain, beta-D-arabinofuranose-(1-2)-alpha-D-arabinofuranose-(1-3)-[beta-D-arabinofuranose-(1-2)-alpha-D-arabinofuranose-(1-5)]alpha-D-arabinofuranose-(1-5)-methyl alpha-D-arabinofuranoside | | Authors: | Murase, T, Zheng, R.B, Joe, M, Bai, Y, Marcus, S.L, Lowary, T.L, Ng, K.K.S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into antibody recognition of mycobacterial polysaccharides.
J.Mol.Biol., 392, 2009
|
|
4NC0
 
 | | Crystal Structure of TcdA-A2 Bound to A26.8 VHH | | Descriptor: | A26.8 VHH, Cell wall-binding repeat protein | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
3H5X
 
 | | Crystal Structure of 2'-amino-2'-deoxy-cytidine-5'-triphosphate bound to Norovirus GII RNA polymerase | | Descriptor: | 2'-amino-2'-deoxycytidine 5'-(tetrahydrogen triphosphate), 5'-R(*UP*GP*CP*CP*CP*GP*GP*G)-3', 5'-R(P*UP*GP*CP*CP*CP*GP*GP*GP*C)-3', ... | | Authors: | Zamyatkin, D.F, Parra, F, Machin, A, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Binding of 2'-amino-2'-deoxycytidine-5'-triphosphate to norovirus polymerase induces rearrangement of the active site.
J.Mol.Biol., 390, 2009
|
|
3H5Y
 
 | | Norovirus polymerase+primer/template+CTP complex at 6 mM MnCl2 | | Descriptor: | 5'-R(*UP*GP*CP*CP*CP*GP*GP*G)-3', 5'-R(P*UP*GP*CP*CP*CP*GP*GP*GP*C)-3', CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Zamyatkin, D.F, Parra, F, Machin, A, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Binding of 2'-amino-2'-deoxycytidine-5'-triphosphate to norovirus polymerase induces rearrangement of the active site.
J.Mol.Biol., 390, 2009
|
|
4NBZ
 
 | | Crystal Structure of TcdA-A1 Bound to A26.8 VHH | | Descriptor: | A26.8 VHH, TcdA | | Authors: | Murase, T, Eugenio, L, Schorr, M, Hussack, G, Tanha, J, Kitova, E.N, Klassen, J.S, Ng, K.K.S. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Antibody Recognition in the Receptor-binding Domains of Toxins A and B from Clostridium difficile.
J.Biol.Chem., 289, 2014
|
|
4QPX
 
 | | NV polymerase post-incorporation-like complex | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Polyprotein, ... | | Authors: | Zamyatkin, D.F, Parra, F, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2014-06-25 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of a backtracked state reveals conformational changes similar to the state following nucleotide incorporation in human norovirus polymerase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
