3LSZ
 
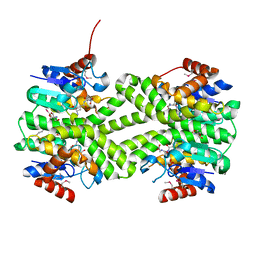 | | Crystal structure of glutathione s-transferase from Rhodobacter sphaeroides | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTATHIONE, GLYCEROL, ... | | Authors: | Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-14 | | Release date: | 2010-03-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glutathione s-transferase from Rhodobacter sphaeroides
To be Published
|
|
3LY0
 
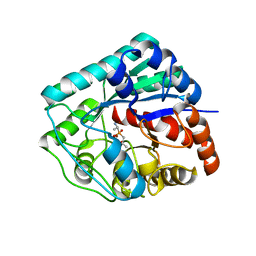 | | Crystal structure of metallo peptidase from Rhodobacter sphaeroides liganded with phosphinate mimic of dipeptide L-Ala-D-Ala | | Descriptor: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, Dipeptidase AC. Metallo peptidase. MEROPS family M19, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Crystal structure of metallo peptidase from Rhodobacter sphaeroides
liganded with phosphinate mimic of dipeptide L-Ala-D-Ala
To be Published
|
|
3LYB
 
 | |
3MQT
 
 | |
3LL3
 
 | |
3LOC
 
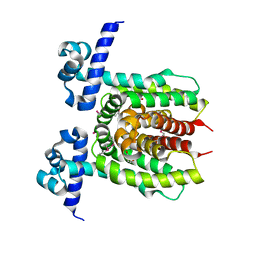 | | Crystal structure of putative transcriptional regulator ycdc | | Descriptor: | HTH-type transcriptional regulator rutR, URACIL | | Authors: | Patskovsky, Y.V, Knapik, A.A, Mennella, V, Burley, S.K, Minor, W, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-03 | | Release date: | 2010-03-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Hypothetical Transcriptional Regulator Ycdc from Escherichia Coli
To be Published
|
|
3ME7
 
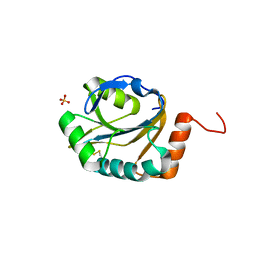 | | Crystal structure of putative electron transport protein aq_2194 from Aquifex aeolicus VF5 | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative electron transport protein aq_2194 from Aquifex aeolicus VF5
To be Published
|
|
3M2T
 
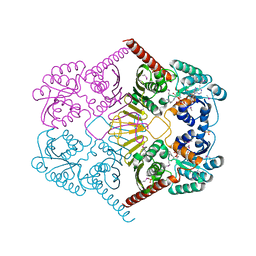 | |
3M3M
 
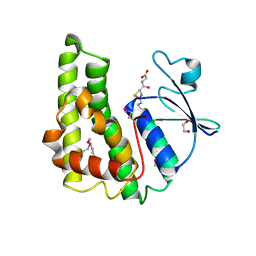 | | Crystal structure of glutathione S-transferase from Pseudomonas fluorescens [Pf-5] | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase, ... | | Authors: | Bagaria, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of glutathione S-transferase from Pseudomonas fluorescens [Pf-5]
To be Published
|
|
3LXZ
 
 | |
3LX6
 
 | |
3LYK
 
 | |
3LYP
 
 | |
3ME8
 
 | | Crystal structure of putative electron transfer protein aq_2194 from Aquifex aeolicus VF5 | | Descriptor: | Putative uncharacterized protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative electron transfer protein aq_2194 from Aquifex aeolicus VF5
To be Published
|
|
3MDN
 
 | |
3MDK
 
 | |
3LUA
 
 | |
3MGG
 
 | |
3MKC
 
 | |
4Q9T
 
 | |
1FI4
 
 | | THE X-RAY CRYSTAL STRUCTURE OF MEVALONATE 5-DIPHOSPHATE DECARBOXYLASE AT 2.3 ANGSTROM RESOLUTION. | | Descriptor: | MEVALONATE 5-DIPHOSPHATE DECARBOXYLASE | | Authors: | Bonanno, J.B, Edo, C, Eswar, N, Pieper, U, Romanowski, M.J, Ilyin, V, Gerchman, S.E, Kycia, H, Studier, F.W, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-08-03 | | Release date: | 2001-03-21 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural genomics of enzymes involved in sterol/isoprenoid biosynthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1F89
 
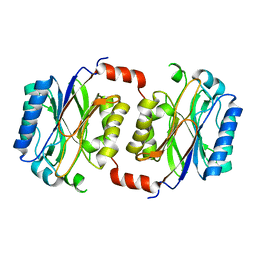 | | Crystal structure of Saccharomyces cerevisiae Nit3, a member of branch 10 of the nitrilase superfamily | | Descriptor: | 32.5 KDA PROTEIN YLR351C | | Authors: | Kumaran, D, Eswaramoorthy, S, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-06-29 | | Release date: | 2001-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative CN hydrolase from yeast
Proteins, 52, 2003
|
|
1HQZ
 
 | | Cofilin homology domain of a yeast actin-binding protein ABP1P | | Descriptor: | ACTIN-BINDING PROTEIN | | Authors: | Strokopytov, B.V, Fedorov, A.A, Mahoney, N, Drubin, D.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-12-20 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phased translation function revisited: structure solution of the cofilin-homology domain from yeast actin-binding protein 1 using six-dimensional searches.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1G61
 
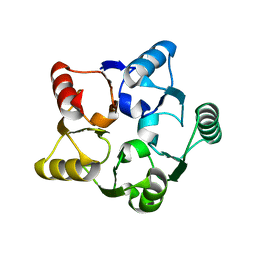 | | CRYSTAL STRUCTURE OF M.JANNASCHII EIF6 | | Descriptor: | TRANSLATION INITIATION FACTOR 6 | | Authors: | Groft, C.M, Beckmann, R, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of ribosome anti-association factor IF6.
Nat.Struct.Biol., 7, 2000
|
|
1G62
 
 | | CRYSTAL STRUCTURE OF S.CEREVISIAE EIF6 | | Descriptor: | RIBOSOME ANTI-ASSOCIATION FACTOR EIF6 | | Authors: | Groft, C.M, Beckmann, R, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of ribosome anti-association factor IF6.
Nat.Struct.Biol., 7, 2000
|
|
