5BQF
 
 | | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADP, HEPES and L(+)-tartaric acid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Langner, K.M, Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Stead, M, Hillerich, B.S, Chowdhury, S, Hammonds, J, Zimmerman, M.D, Al Obaidi, N, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADP, HEPES and L-tartaric acid
to be published
|
|
4MDC
 
 | | Crystal structure of glutathione S-transferase from Sinorhizobium meliloti 1021, NYSGRC target 021389 | | Descriptor: | GLYCEROL, Putative glutathione S-transferase | | Authors: | Shabalin, I.G, Bacal, P, Cooper, D.R, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-22 | | Release date: | 2013-09-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of glutathione S-transferase from Sinorhizobium meliloti 1021
To be Published
|
|
3DEC
 
 | | Crystal structure of a glycosyl hydrolases family 2 protein from Bacteroides thetaiotaomicron | | Descriptor: | Beta-galactosidase, POTASSIUM ION | | Authors: | Kumaran, D, Bonanno, J, Romero, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-09 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Glycosyl Hydrolases Family 2 protein from Bacteroides thetaiotaomicron.
To be Published
|
|
4XK2
 
 | | Crystal structure of aldo-keto reductase from Polaromonas sp. JS666 | | Descriptor: | Aldo/keto reductase, CHLORIDE ION, SODIUM ION | | Authors: | Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Sroka, P, Hillerich, B.S, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-01-09 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of aldo-keto reductase from Polaromonas sp. JS666
to be published
|
|
3RP1
 
 | | Crystal structure of Human LAIR-1 in C2 space group | | Descriptor: | Leukocyte-associated immunoglobulin-like receptor 1 | | Authors: | Sampathkumar, P, Ramagopal, U.A, Yan, Q, Toro, R, Nathenson, S, Bonanno, J, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-26 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Human LAIR-1 in C2 space group
To be Published
|
|
4JGJ
 
 | | Crystal structure of the Ig-like D1 domain from mouse Carcinoembryogenic antigen-related cell adhesion molecule 15 (CEACAM15) [PSI-NYSGRC-005691] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 15, Unknown peptide | | Authors: | Kumar, P.R, Bonanno, J, Ahmed, M, Banu, R, Bhosle, R, Calarese, D, Celikigil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-03-01 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6508 Å) | | Cite: | Crystal structure of the Ig-like D1 domain of CEACAM15 from Mus musculus [NYSGRC-005691]
to be published
|
|
3FEQ
 
 | | Crystal structure of uncharacterized protein eah89906 | | Descriptor: | PUTATIVE AMIDOHYDROLASE, ZINC ION | | Authors: | Patskovsky, Y, Bonanno, J, Romero, R, Freeman, J, Lau, C, Smith, D, Bain, K, Wasserman, S.R, Raushel, F, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-30 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
4OO6
 
 | | Crystal structure of human KAP-beta2 bound to the NLS of HCC1 (Hepato Cellular Carcinoma protein 1) | | Descriptor: | RNA-binding protein 39, Transportin-1 | | Authors: | Sampathkumar, P, Brower, A, Soniat, M, Bonanno, J, Hillerich, B, Seidel, R.D, Rout, M.P, Chook, Y.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2014-01-30 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human KAP-beta2 bound to the NLS of HCC1 (Hepato Cellular Carcinoma protein 1)
to be published
|
|
4PMJ
 
 | | Crystal structure of a putative oxidoreductase from Sinorhizobium meliloti 1021 in complex with NADP | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Szlachta, K, Zimmerman, M.D, Hillerich, B.S, Gizzi, A, Toro, R, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-05-21 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a putative oxidoreductase from Sinorhizobiummeliloti 1021 in complex with NADP
to be published
|
|
4MI5
 
 | | Crystal structure of the EZH2 SET domain | | Descriptor: | Histone-lysine N-methyltransferase EZH2, SULFATE ION, ZINC ION | | Authors: | Antonysamy, S, Condon, B, Druzina, Z, Bonanno, J, Gheyi, T, Macewan, I, Zhang, A, Ashok, S, Russell, M, Luz, J.G. | | Deposit date: | 2013-08-30 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Context of Disease-Associated Mutations and Putative Mechanism of Autoinhibition Revealed by X-Ray Crystallographic Analysis of the EZH2-SET Domain.
Plos One, 8, 2013
|
|
6NG3
 
 | | Crystal structure of human CD160 and HVEM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen,Tumor necrosis factor receptor superfamily member 14, MAGNESIUM ION, ... | | Authors: | Liu, W, Bonanno, J, Almo, S.C. | | Deposit date: | 2018-12-21 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Basis of CD160:HVEM Recognition.
Structure, 27, 2019
|
|
6NG9
 
 | |
6NGG
 
 | |
3ZH3
 
 | |
4JJH
 
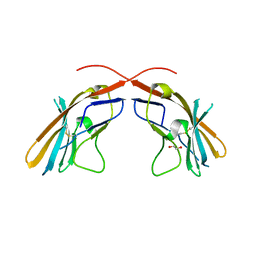 | | Crystal structure of the D1 domain from human Nectin-4 extracellular fragment [PSI-NYSGRC-005624] | | Descriptor: | 1,2-ETHANEDIOL, Poliovirus receptor-related protein 4 | | Authors: | Kumar, P.R, Bonanno, J, Ahmed, M, Banu, R, Bhosle, R, Calarese, D, Celikigil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-03-07 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the D1 domain of human Nectin-4 extracellular fragment [NYSGRC-005624]
to be published
|
|
4JYL
 
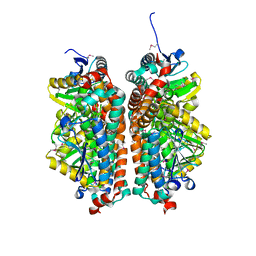 | | Crystal structure of enoyl-CoA hydratase from Thermoplasma volcanium GSS1 | | Descriptor: | CHLORIDE ION, Enoyl-CoA hydratase, SULFATE ION | | Authors: | Shabalin, I.G, Cooper, D.R, Majorek, K.A, Mikolajczak, K, Porebski, P.J, Stead, M, Hillerich, B.S, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-17 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of enoyl-CoA hydratase from Thermoplasma volcanium GSS1
To be Published
|
|
5VE2
 
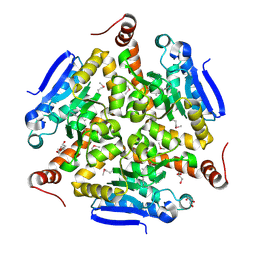 | | Crystal structure of enoyl-CoA hydratase/isomerase from Pseudoalteromonas atlantica T6c at 2.3 A resolution. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase, GLYCEROL, ... | | Authors: | Siuda, M.K, Shabalin, I.G, Cooper, D.R, Chapman, H.C, Tkaczuk, K.L, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-04-03 | | Release date: | 2017-04-19 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of enoyl-CoA hydratase/isomerase from Pseudoalteromonas atlantica T6c at 2.3 A resolution.
to be published
|
|
2QDD
 
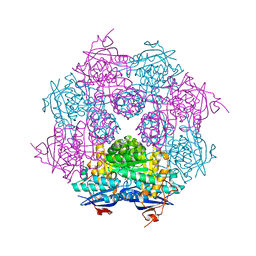 | | Crystal structure of a member of enolase superfamily from Roseovarius nubinhibens ISM | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Patskovsky, Y, Bonanno, J, Sauder, J.M, Gilmore, J.M, Iizuka, M, Groshong, C, Gheyi, T, Sojitra, S, Wasserman, S.R, Koss, J, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-06-20 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a member of enolase superfamily from Roseovarius nubinhibens ISM.
To be Published
|
|
2QSD
 
 | | Crystal structure of a protein Il1583 from Idiomarina loihiensis | | Descriptor: | GLYCEROL, Uncharacterized conserved protein | | Authors: | Patskovsky, Y, Bonanno, J, Sauder, J.M, Romero, R, Rutter, M, Koss, J, Mckenzie, C, Gheyi, T, Bain, K, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-14 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Protein Il1583 from Idiomarina loihiensis.
To be Published
|
|
7SGS
 
 | | Cryo-EM structure of full-length MAP7 bound to the microtubule | | Descriptor: | Ensconsin, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ferro, L.S, Fang, Q, Eshun-Wilson, L, Fernandes, J, Jack, A, Farrell, D.P, Golcuk, M, Huijben, T, Costa, K, Gur, M, DiMaio, F, Nogales, E, Yildiz, A. | | Deposit date: | 2021-10-07 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional insight into regulation of kinesin-1 by microtubule-associated protein MAP7.
Science, 375, 2022
|
|
4N1S
 
 | |
4N1O
 
 | |
4D8L
 
 | | Crystal structure of the 2-pyrone-4,6-dicarboxylic acid hydrolase from sphingomonas paucimobilis | | Descriptor: | 2-pyrone-4,6-dicarbaxylate hydrolase | | Authors: | Malashkevich, V.N, Toro, R, Bonanno, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2012-01-10 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
4N1N
 
 | |
4N1P
 
 | |
