5K7M
 
 | |
5K7W
 
 | |
3DIN
 
 | | Crystal structure of the protein-translocation complex formed by the SecY channel and the SecA ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Zimmer, J, Nam, Y, Rapoport, T.A. | | Deposit date: | 2008-06-20 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of a complex of the ATPase SecA and the protein-translocation channel.
Nature, 455, 2008
|
|
7RX7
 
 | |
3PE4
 
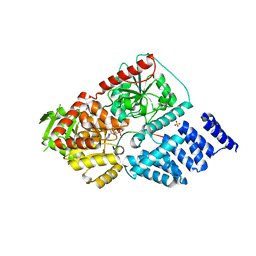 | | Structure of human O-GlcNAc transferase and its complex with a peptide substrate | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, ... | | Authors: | Lazarus, M.B, Nam, Y, Jiang, J, Sliz, P, Walker, S. | | Deposit date: | 2010-10-25 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of human O-GlcNAc transferase and its complex with a peptide substrate.
Nature, 469, 2011
|
|
3PE3
 
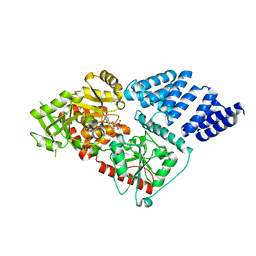 | | Structure of human O-GlcNAc transferase and its complex with a peptide substrate | | Descriptor: | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, URIDINE-5'-DIPHOSPHATE | | Authors: | Lazarus, M.B, Nam, Y, Jiang, J, Sliz, P, Walker, S. | | Deposit date: | 2010-10-25 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure of human O-GlcNAc transferase and its complex with a peptide substrate.
Nature, 469, 2011
|
|
6DU5
 
 | |
4PDT
 
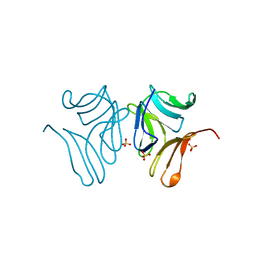 | | Japanese Marasmius oreades lectin | | Descriptor: | Mannose recognizing lectin, SULFATE ION | | Authors: | Noma, Y, Shimokawa, M, Maeganeku, C, Motoshima, H, Watanabe, K, Minami, Y, Yagi, F. | | Deposit date: | 2014-04-22 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of Japanese Marasmius oreades lectin at 1.40 Angstroms resolution.
To Be Published
|
|
8EG0
 
 | |
8D9K
 
 | |
8D9L
 
 | |
8D59
 
 | | Crystal structure of human METTL1 in complex with SAM | | Descriptor: | CACODYLATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raj, R, Babu, K, Nam, Y. | | Deposit date: | 2022-06-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures and mechanisms of tRNA methylation by METTL1-WDR4.
Nature, 613, 2023
|
|
8D5B
 
 | | Crystal structure of human METTL1 in complex with SAH | | Descriptor: | CACODYLATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raj, R, Babu, K, Nam, Y. | | Deposit date: | 2022-06-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures and mechanisms of tRNA methylation by METTL1-WDR4.
Nature, 613, 2023
|
|
8D58
 
 | | Crystal structure of human METTL1-WDR4 complex | | Descriptor: | CHLORIDE ION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Raj, R, Babu, K, Nam, Y. | | Deposit date: | 2022-06-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structures and mechanisms of tRNA methylation by METTL1-WDR4.
Nature, 613, 2023
|
|
4TKC
 
 | | Japanese Marasmius oreades lectin complexed with mannose | | Descriptor: | GLYCEROL, Mannose recognizing lectin, alpha-D-mannopyranose, ... | | Authors: | Noma, Y, Shimokawa, M, Maeganeku, C, Motoshima, H, Watanabe, K, Minami, Y, Yagi, F. | | Deposit date: | 2014-05-26 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structure of Japanese Marasmius oreades lectin complexed with mannose.
To Be Published
|
|
3WTQ
 
 | | Crystal structure of VDR-LBD complexed with 22S-butyl-2-methylidene-19-nor-1a,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3S)-3-butyl-6-hydroxy-6-methylheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Inaba, Y, Itoh, T, Anami, Y, Ikura, T, Ito, N, Yamamoto, K. | | Deposit date: | 2014-04-15 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of VDR-LBD complexed with 22S-butyl-2-methylidene-19-nor-1a,25-dihydroxyvitamin D3
To be Published
|
|
2YZQ
 
 | | Crystal structure of uncharacterized conserved protein from Pyrococcus horikoshii | | Descriptor: | Putative uncharacterized protein PH1780, S-ADENOSYLMETHIONINE | | Authors: | Kanagawa, M, Minami, Y, Watanabe, N, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of uncharacterized conserved protein from Pyrococcus horikoshii
To be Published
|
|
5H1E
 
 | | Interaction between vitamin D receptor and coactivator peptide SRC2-3 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Nuclear receptor coactivator 2 peptide, Vitamin D3 receptor | | Authors: | Egawa, D, Itoh, T, Kato, A, Kataoka, S, Anami, Y, Yamamoto, K. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | SRC2-3 binds to vitamin D receptor with high sensitivity and strong affinity
Bioorg. Med. Chem., 25, 2017
|
|
7C4N
 
 | | Ancestral L-amino acid oxidase (AncLAAO-N5) L-Phe binding form | | Descriptor: | Ancestral L-amino acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, PHENYLALANINE | | Authors: | Nakano, S, Minamino, Y, Karasuda, H, Ito, S. | | Deposit date: | 2020-05-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ancestral L-amino acid oxidases for deracemization and stereoinversion of amino acids
Commun Chem, 3, 2020
|
|
7C4L
 
 | | Anncestral L-amino acid oxidase (AncLAAO-N5) L-Gln binding form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTAMINE, L-amino acid oxidase | | Authors: | Nakano, S, Minamino, Y, Karasuda, H, Ito, S. | | Deposit date: | 2020-05-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ancestral L-amino acid oxidases for deracemization and stereoinversion of amino acids
Commun Chem, 3, 2020
|
|
7C4M
 
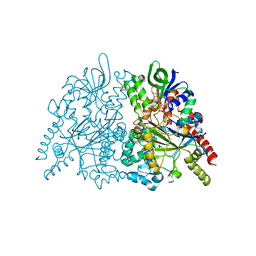 | | Ancestral L-amino acid oxidase (AncLAAO-N5) L-Trp binding form | | Descriptor: | Ancestral L-amino acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, TRYPTOPHAN | | Authors: | Nakano, S, Minamino, Y, Karasuda, H, Ito, S. | | Deposit date: | 2020-05-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ancestral L-amino acid oxidases for deracemization and stereoinversion of amino acids
Commun Chem, 3, 2020
|
|
7C4K
 
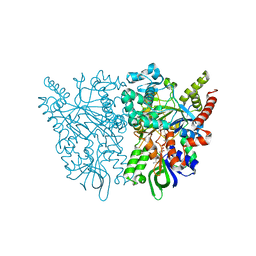 | | Ancestral L-amino acid oxidase (AncLAAO-N5) ligand free form | | Descriptor: | Ancestral L-amino acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Nakano, S, Minamino, Y, Karasuda, H, Ito, S. | | Deposit date: | 2020-05-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ancestral L-amino acid oxidases for deracemization and stereoinversion of amino acids
Commun Chem, 3, 2020
|
|
5ZWI
 
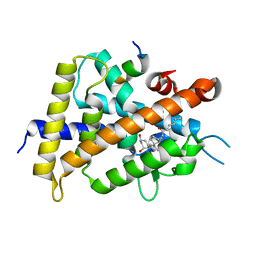 | | Interaction between Vitamin D receptor (VDR) and a ligand having a dienone group | | Descriptor: | (2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-4,6-diene-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, Vitamin D3 receptor | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
5ZWH
 
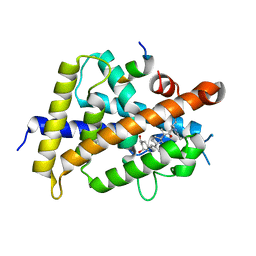 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having an ene-ynone group via conjugate addition reaction | | Descriptor: | (2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-4,6-diene-3-one, (E,2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-6-en-4-yn-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, ... | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
2D2A
 
 | | Crystal Structure of Escherichia coli SufA Involved in Biosynthesis of Iron-sulfur Clusters | | Descriptor: | SufA protein | | Authors: | Wada, K, Hasegawa, Y, Gong, Z, Minami, Y, Fukuyama, K, Takahashi, Y. | | Deposit date: | 2005-09-05 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Escherichia coli SufA involved in biosynthesis of iron-sulfur clusters: Implications for a functional dimer
Febs Lett., 579, 2005
|
|
