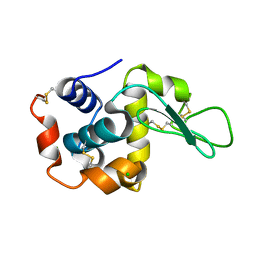
6JXQ
 
 | |
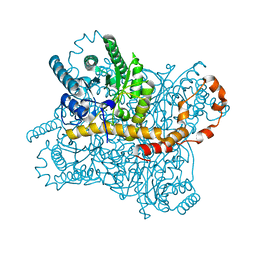
6KD2
 
 | |
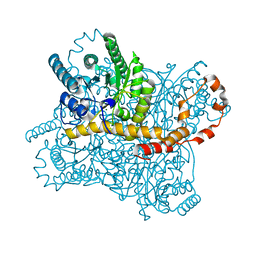
6KCA
 
 | |
6JXP
 
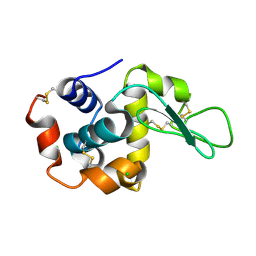 | |
6KD1
 
 | |
6K1X
 
 | |
6KCD
 
 | |
6KCB
 
 | |
6K1W
 
 | |
6KCC
 
 | |
6K1Y
 
 | |
7WKR
 
 | |
7WUC
 
 | |
7WBE
 
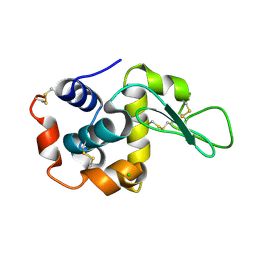 | |
7WBD
 
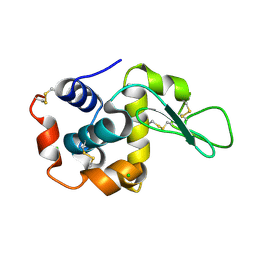 | |
7WBF
 
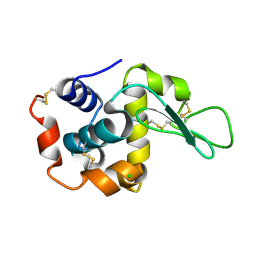 | | Crystal structure of lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Nam, K.H. | | Deposit date: | 2021-12-16 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Processing of Multicrystal Diffraction Patterns in Macromolecular Crystallography Using Serial Crystallography Programs.
Crystals, 12, 2022
|
|
7XF7
 
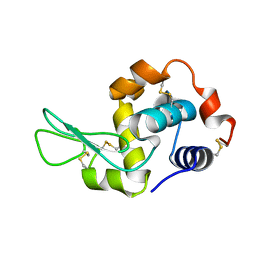 | |
7XF6
 
 | | Crystal Structure of Human Lysozyme | | Descriptor: | ACETATE ION, Lysozyme C | | Authors: | Nam, K.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Human Lysozyme Complexed with N-Acetyl-alpha-d-glucosamine.
Appl Sci (Basel), 12, 2022
|
|
7XF8
 
 | |
6LNG
 
 | | Rapid crystallization of streptavidin using charged peptides | | Descriptor: | GLYCEROL, Streptavidin | | Authors: | Minamihata, K, Tsukamoto, K, Adachi, M, Shimizu, R, Mishina, M, Kuroki, R, Nagamune, T. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8000015 Å) | | Cite: | Genetically fused charged peptides induce rapid crystallization of proteins.
Chem.Commun.(Camb.), 56, 2020
|
|
5L6Q
 
 | | Refolded AL protein from cardiac amyloidosis | | Descriptor: | CARBONATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Annamalai, K, Liberta, F, Vielberg, M.-T, Lilie, H, Guehrs, K.-H, Schierhorn, A, Koehler, R, Schmidt, A, Haupt, C, Hegenbart, O, Schoenland, S, Groll, M, Faendrich, M. | | Deposit date: | 2016-05-31 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Common Fibril Structures Imply Systemically Conserved Protein Misfolding Pathways In Vivo.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6GV1
 
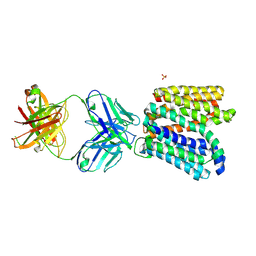 | | Crystal structure of E.coli Multidrug/H+ antiporter MdfA in outward open conformation with bound Fab fragment | | Descriptor: | Fab fragment YN1074 heavy chain, Fab fragment YN1074 light chain, Major Facilitator Superfamily multidrug/H+ antiporter MdfA from E.coli, ... | | Authors: | Nagarathinam, K, Parthier, C, Stubbs, M.T, Tanabe, M. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Outward open conformation of a Major Facilitator Superfamily multidrug/H+antiporter provides insights into switching mechanism.
Nat Commun, 9, 2018
|
|
1G91
 
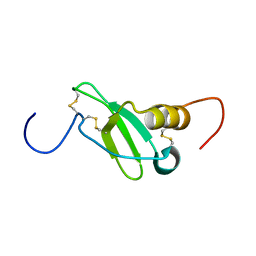 | | SOLUTION STRUCTURE OF MYELOID PROGENITOR INHIBITORY FACTOR-1 (MPIF-1) | | Descriptor: | MYELOID PROGENITOR INHIBITORY FACTOR-1 | | Authors: | Rajarathnam, K, Li, Y, Rohrer, T, Gentz, R. | | Deposit date: | 2000-11-21 | | Release date: | 2001-03-07 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of myeloid progenitor inhibitory factor-1 (MPIF-1), a novel monomeric CC chemokine.
J.Biol.Chem., 276, 2001
|
|
7KKF
 
 | |
6IAK
 
 | | The crystal structure of the chicken CREB3 bZIP | | Descriptor: | Uncharacterized protein | | Authors: | Sabaratnam, K, Renner, M. | | Deposit date: | 2018-11-26 | | Release date: | 2019-12-11 | | Last modified: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Insights from the crystal structure of the chicken CREB3 bZIP suggest that members of the CREB3 subfamily transcription factors may be activated in response to oxidative stress.
Protein Sci., 28, 2019
|
|
