2AI5
 
 | | Solution Structure of Cytochrome C552, determined by Distributed Computing Implementation for NMR data | | 分子名称: | Cytochrome c-552, HEME C | | 著者 | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | 登録日 | 2005-07-29 | | 公開日 | 2006-05-23 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
8IUA
 
 | |
8IU9
 
 | |
8IU8
 
 | |
8IUC
 
 | |
8IUB
 
 | |
6AKG
 
 | |
6AKE
 
 | |
6AKF
 
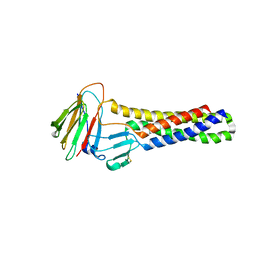 | |
7C95
 
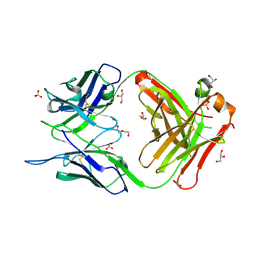 | | Crystal structure of the anti-human podoplanin antibody Fab fragment | | 分子名称: | GLYCEROL, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | 著者 | Nakamura, S, Suzuki, K, Ogasawara, S, Naruchi, K, Shimabukuro, J, Tukahara, N, Kaneko, M.K, Kato, Y, Murata, T. | | 登録日 | 2020-06-04 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Crystal structure of an anti-podoplanin antibody bound to a disialylated O-linked glycopeptide.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
2DKN
 
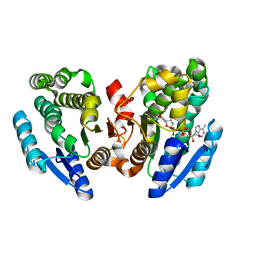 | |
2D0S
 
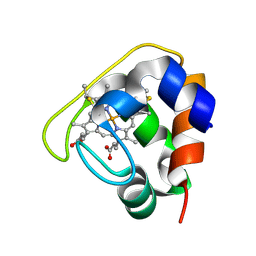 | | Crystal structure of the Cytochrome C552 from moderate thermophilic bacterium, hydrogenophilus thermoluteolus | | 分子名称: | HEME C, cytochrome c | | 著者 | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | 登録日 | 2005-08-08 | | 公開日 | 2006-05-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
2Z2T
 
 | |
7FE3
 
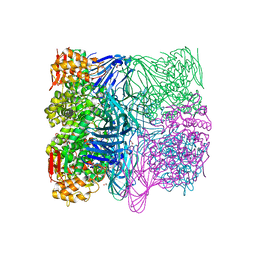 | |
7FE4
 
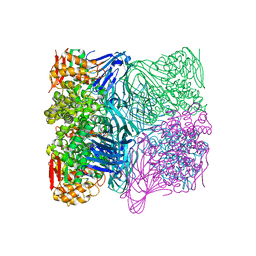 | |
8XY0
 
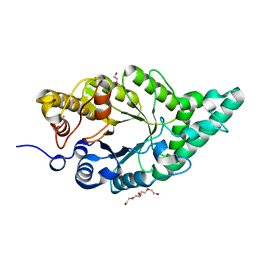 | | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR | | 分子名称: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Endo-1,4-beta-xylanase A | | 著者 | Nakamura, T, Takita, T, Mizutani, K, Mikami, B, Nakamura, S, Yasukawa, K. | | 登録日 | 2024-01-19 | | 公開日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR.
Sci Rep, 14, 2024
|
|
8Y1M
 
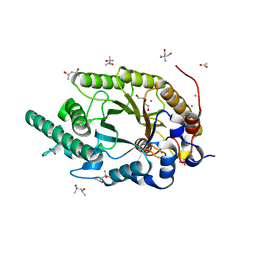 | | Xylanase R from Bacillus sp. TAR-1 complexed with xylobiose. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | 著者 | Nakamura, T, Kuwata, K, Takita, T, Mizutani, K, Mikami, B, Nakamura, S, Yasukawa, K. | | 登録日 | 2024-01-25 | | 公開日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Activity-stability trade-off observed in variants at position 315 of the GH10 xylanase XynR.
Sci Rep, 14, 2024
|
|
2HYK
 
 | | The crystal structure of an endo-beta-1,3-glucanase from alkaliphilic Nocardiopsis sp.strain F96 | | 分子名称: | Beta-1,3-glucanase, CALCIUM ION, ETHANOL, ... | | 著者 | Fibriansah, G, Nakamura, S, Kumasaka, T. | | 登録日 | 2006-08-07 | | 公開日 | 2007-10-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | The 1.3 A crystal structure of a novel endo-beta-1,3-glucanase of glycoside hydrolase family 16 from alkaliphilic Nocardiopsis sp. strain F96.
Proteins, 69, 2007
|
|
4U4P
 
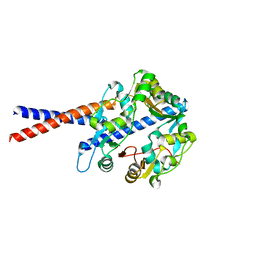 | | Crystal structure of the human condensin SMC hinge domain heterodimer with short coiled coils | | 分子名称: | Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | 著者 | Uchiyama, S, Kawahara, K, Hosokawa, Y, Fukakusa, S, Oki, H, Nakamura, S, Noda, M, Takino, R, Miyahara, Y, Maruno, T, Kobayashi, Y, Ohkubo, T, Fukui, K. | | 登録日 | 2014-07-24 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structural basis for dimer information and DNA recognition of human SMC proteins
to be published
|
|
8IU0
 
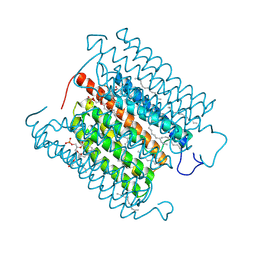 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc | | 分子名称: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | 著者 | Tajima, S, Kim, Y, Nakamura, S, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | 登録日 | 2023-03-23 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
1WZB
 
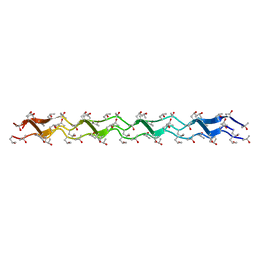 | | Crystal structure of the collagen triple helix model [{HYP(R)-HYP(R)-GLY}10]3 | | 分子名称: | Collagen triple helix | | 著者 | Kawahara, K, Nakamura, S, Nishi, Y, Uchiyama, S, Nishiuchi, Y, Nakazawa, T, Ohkubo, T, Kobayashi, Y. | | 登録日 | 2005-03-03 | | 公開日 | 2006-01-31 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Effect of hydration on the stability of the collagen-like triple-helical structure of [4(R)-hydroxyprolyl-4(R)-hydroxyprolylglycine]10
Biochemistry, 44, 2005
|
|
1YT6
 
 | | NMR structure of peptide SD | | 分子名称: | peptide SD | | 著者 | Murata, T, Hemmi, H, Nakamura, S, Shimizu, K, Suzuki, Y, Yamaguchi, I. | | 登録日 | 2005-02-10 | | 公開日 | 2005-09-27 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, epitope mapping, and docking simulation of a gibberellin mimic peptide as a peptidyl mimotope for a hydrophobic ligand.
Febs J., 272, 2005
|
|
5AX6
 
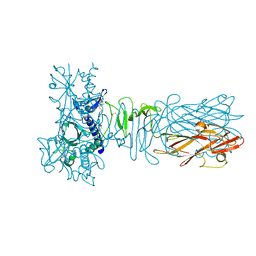 | | The crystal structure of CofB, the minor pilin subunit of CFA/III from human enterotoxigenic Escherichia coli. | | 分子名称: | ACETATE ION, CofB | | 著者 | Kawahara, K, Oki, K, Fukaksua, F, Maruno, T, Kobayashi, Y, Daisuke, M, Taniguchi, T, Honda, T, Iida, T, Nakamura, S, Ohkubo, T. | | 登録日 | 2015-07-16 | | 公開日 | 2016-03-09 | | 最終更新日 | 2020-02-26 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Homo-trimeric Structure of the Type IVb Minor Pilin CofB Suggests Mechanism of CFA/III Pilus Assembly in Human Enterotoxigenic Escherichia coli
J.Mol.Biol., 428, 2016
|
|
8WG1
 
 | | Crystal structure of GH97 glucodextranase mutant E509Q from Flavobacterium johnsoniae in complex with panose | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Candidate alpha-glucosidase Glycoside hydrolase family 97, ... | | 著者 | Kurata, R, Nakamura, S, Miyazaki, T. | | 登録日 | 2023-09-20 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structural insights into alpha-(1→6)-linkage preference of GH97 glucodextranase from Flavobacterium johnsoniae.
Febs J., 291, 2024
|
|
8WG0
 
 | | Crystal structure of GH97 glucodextranase from Flavobacterium johnsoniae in complex with glucose | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Candidate alpha-glucosidase Glycoside hydrolase family 97, ... | | 著者 | Kurata, R, Nakamura, S, Miyazaki, T. | | 登録日 | 2023-09-20 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural insights into alpha-(1→6)-linkage preference of GH97 glucodextranase from Flavobacterium johnsoniae.
Febs J., 291, 2024
|
|
