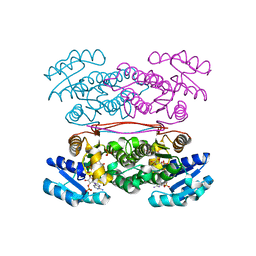
3ASU
 
 | |
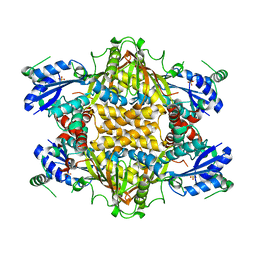
1KH1
 
 | |
2DZD
 
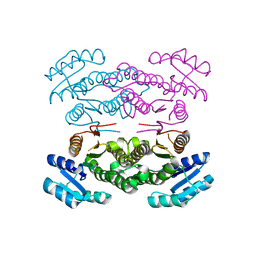
 | | Crystal structure of the biotin carboxylase domain of pyruvate carboxylase | | Descriptor: | pyruvate carboxylase | | Authors: | Kondo, S, Nakajima, Y, Sugio, S, Sueda, S, Islam, M.N, Kondo, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the biotin carboxylase domain of pyruvate carboxylase from Bacillus thermodenitrificans
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
3ASV
 
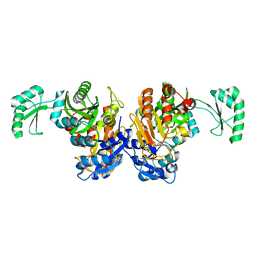
 | | The Closed form of serine dehydrogenase complexed with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PHOSPHATE ION, Short-chain dehydrogenase/reductase SDR | | Authors: | Yamazawa, R, Nakajima, Y, Yoshimoto, T, Ito, K. | | Deposit date: | 2010-12-21 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of serine dehydrogenase from Escherichia coli: important role of the C-terminal region for closed-complex formation.
J.Biochem., 149, 2011
|
|
1WMB
 
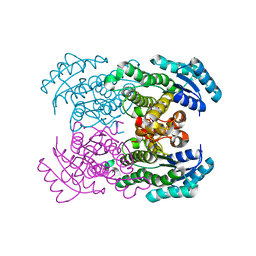 | | Crystal structure of NAD dependent D-3-hydroxybutylate dehydrogenase | | Descriptor: | CACODYLATE ION, D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION | | Authors: | Ito, K, Nakajima, Y, Ichihara, E, Ogawa, K, Yoshimoto, T. | | Deposit date: | 2004-07-06 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | d-3-Hydroxybutyrate Dehydrogenase from Pseudomonas fragi: Molecular Cloning of the Enzyme Gene and Crystal Structure of the Enzyme
J.Mol.Biol., 355, 2006
|
|
2ZTM
 
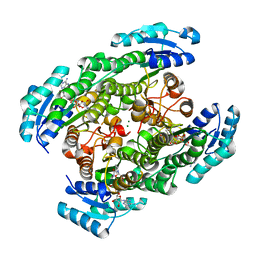 | | T190S mutant of D-3-hydroxybutyrate dehydrogenase | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-07 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
2ZTL
 
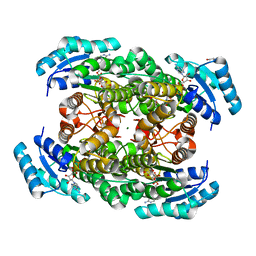 | | Closed conformation of D-3-hydroxybutyrate dehydrogenase complexed with NAD+ and L-3-hydroxybutyrate | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, D(-)-3-hydroxybutyrate dehydrogenase, GLYCEROL, ... | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-07 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
2ZTU
 
 | | T190A mutant of D-3-hydroxybutyrate dehydrogenase complexed with NAD+ | | Descriptor: | D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-09 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
2ZTV
 
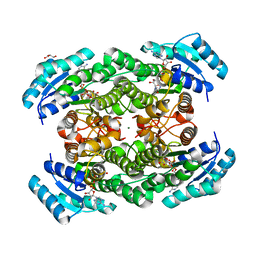 | | The binary complex of D-3-hydroxybutyrate dehydrogenase with NAD+ | | Descriptor: | D(-)-3-hydroxybutyrate dehydrogenase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-09 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
1X3X
 
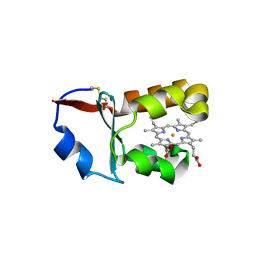 | | Crystal Structure of Cytochrome b5 from Ascaris suum | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Yokota, T, Nakajima, Y, Yamakura, F, Sugio, S, Hashimoto, M, Takamiya, S, Aoki, T. | | Deposit date: | 2005-05-11 | | Release date: | 2006-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unique structure of Ascaris suum b5-type cytochrome: an additional alpha-helix and positively charged residues on the surface domain interact with redox partners
Biochem.J., 394, 2006
|
|
