1QAI
 
 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*CP*AP*TP*G)-3'), MERCURY (II) ION, REVERSE TRANSCRIPTASE | | Authors: | Najmudin, S, Cote, M, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | Deposit date: | 1999-03-12 | | Release date: | 2000-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain.
J.Mol.Biol., 296, 2000
|
|
7QUZ
 
 | | Crystal structure of the SeMet octameric C-terminal Big_2-CBM56 domains from Paenibacillus illinoisensis (Bacillus circulans IAM1165) beta-1,3-glucanase H | | Descriptor: | Beta-1,3-glucanase bglH, CHLORIDE ION, GLYCEROL | | Authors: | Najmudin, S, Venditto, I, Fontes, C.M.G.A, Bule, P. | | Deposit date: | 2022-01-19 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Structural and biochemical characterization of C-terminal Big_2-CBM56 domains of Bacillus circulans IAM1165 beta-1,3-glucanase H and Paenibacillus sp CBM56
To be published
|
|
1QAJ
 
 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*CP*AP*TP*G)-3'), REVERSE TRANSCRIPTASE | | Authors: | Najmudin, S, Cote, M, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | Deposit date: | 1999-03-18 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain.
J.Mol.Biol., 296, 2000
|
|
1D0E
 
 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | Descriptor: | 5'-D(*TP*TP*TP*CP*AP*TP*GP*CP*AP*TP*G)-3', REVERSE TRANSCRIPTASE | | Authors: | Najmudin, S, Cote, M.L, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | Deposit date: | 1999-09-09 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain
J.Mol.Biol., 296, 2000
|
|
1GCS
 
 | | STRUCTURE OF THE BOVINE GAMMA-B CRYSTALLIN AT 150K | | Descriptor: | GAMMA-B CRYSTALLIN | | Authors: | Najmudin, S, Lindley, P, Slingsby, C, Bateman, O, Myles, D, Kumaraswamy, S, Glover, I. | | Deposit date: | 1994-01-27 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Bovine Gamma-B Crystallin at 150K
J.CHEM.SOC.,FARADAY TRANS., 89, 1993
|
|
8C41
 
 | | High resolution structure of the Streptococcus pneumoniae topoisomerase IV-DNA complex with the novel fluoroquinolone Delafloxacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*AP*TP*GP*AP*AP*T)-3'), ... | | Authors: | Najmudin, S, Pan, X.S, Wang, B, Chayen, N.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | The nature of the molecular interactions at high resolution of the Streptococcus pneumoniae topoisomerase IV-DNA complex with the novel fluoroquinolone Delafloxacin.
To Be Published
|
|
3ERX
 
 | | High-resolution structure of Paracoccus pantotrophus pseudoazurin | | Descriptor: | COPPER (II) ION, Pseudoazurin, SULFATE ION | | Authors: | Najmudin, S, Pauleta, S.R, Moura, I, Romao, M.J. | | Deposit date: | 2008-10-03 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The 1.4 A resolution structure of Paracoccus pantotrophus pseudoazurin.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7PB3
 
 | | Structural and Functional analysis of the Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii | | Descriptor: | PYRROLE-2-CARBOXYLATE, Proline racemase A (AsProR) | | Authors: | Najmudin, S, Pan, X.-S, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Structural and Functional analysis of the Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii
To Be Published
|
|
8QMB
 
 | | Nucleant-assisted 2.0 A resolution structure of the Streptococcus pneumoniae topoisomerase IV-V18mer DNA complex with the novel fluoroquinolone Delafloxacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Najmudin, S, Pan, X.S, Wang, B, Chayen, N.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2023-09-21 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The nature of the molecular interactions at high resolution of the Streptococcus pneumoniae topoisomerase IV-DNA complex with the novel fluoroquinolone Delafloxacin.
To Be Published
|
|
8QMC
 
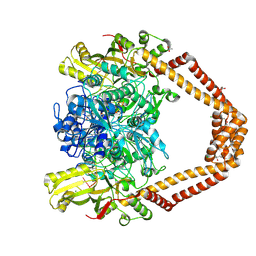 | | High resolution structure of the Streptococcus pneumoniae topoisomerase IV-complex with the V-site 18mer dsDNA and novel fluoroquinolone Delafloxacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Najmudin, S, Pan, X.S, Wang, B, Chayen, N.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2023-09-21 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | The nature of the molecular interactions at high resolution of the Streptococcus pneumoniae topoisomerase IV-DNA complex with the novel fluoroquinolone Delafloxacin.
To Be Published
|
|
7R1N
 
 | | Crystal structure of the Tetrameric C-terminal Big_2-CBM56 domains from Paenibacillus illinoisensis (Bacillus circulans IAM1165) beta-1,3-glucanase H | | Descriptor: | Beta-1,3-glucanase bglH, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Najmudin, S, Venditto, I, Fontes, C.M.G.A, Bule, P. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.072 Å) | | Cite: | Structural and biochemical characterization of C-terminal Big_2-CBM56 domains of Paenibacillus illinoisensis IAM1165 beta-1,3-glucanase H and Paenibacillus sp CBM56
To be published
|
|
7R3T
 
 | | Crystal structure of the Dimeric C-terminal Big_2-CBM56 domains from Paenibacillus illinoisensis (Bacillus circulans IAM1165) beta-1,3-glucanase H | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Beta-1,3-glucanase bglH, CHLORIDE ION, ... | | Authors: | Najmudin, S, Venditto, I, Fontes, C.M.G.A, Bule, P. | | Deposit date: | 2022-02-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Structural and biochemical characterization of C-terminal Big_2-CBM56 domains of Paenibacillus illinoisensis IAM1165 beta-1,3-glucanase H and Paenibacillus sp CBM56
To be published
|
|
2C4X
 
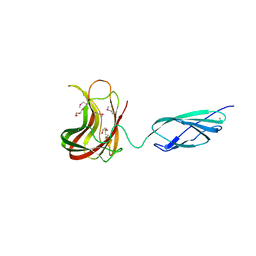 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-10-25 | | Release date: | 2005-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
2C26
 
 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-09-26 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
5LXV
 
 | |
8A4R
 
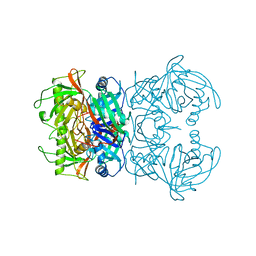 | | Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii from isotropic orthorhombic data at 3.59 A | | Descriptor: | PYRROLE-2-CARBOXYLATE, Proline racemase A (AsProR) | | Authors: | Najmudin, S, Pan, X.-S, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Functional analysis of the Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii
To Be Published
|
|
8A3F
 
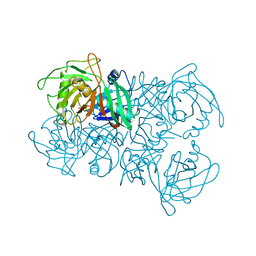 | | Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii from isotropic tetragonal data at 3.15 A | | Descriptor: | PYRROLE-2-CARBOXYLATE, Proline racemase A (AsProR) | | Authors: | Najmudin, S, Pan, X.-S, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2022-06-08 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural and Functional analysis of the Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii
To Be Published
|
|
2W5F
 
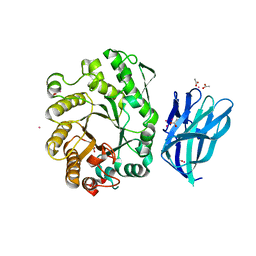 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | Descriptor: | ACETATE ION, CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2008-12-10 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
2WFB
 
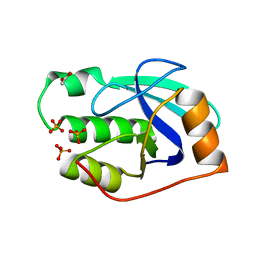 | | High resolution structure of the apo form of the orange protein (ORP) from Desulfovibrio gigas | | Descriptor: | ACETATE ION, PHOSPHATE ION, PUTATIVE UNCHARACTERIZED PROTEIN ORP | | Authors: | Najmudin, S, Bonifacio, C, Duarte, A.G, Pereira, A.S, Moura, I, Moura, J.M, Romao, M.J. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Crystal Structure of the Apo Form of the Orange Protein (Apo-Orp) from Desulfovibrio Gigas
To be Published
|
|
2WZE
 
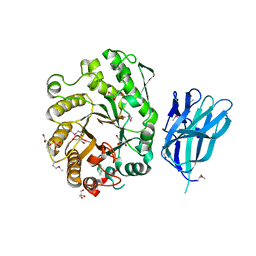 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y, GLYCEROL, ... | | Authors: | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2009-11-27 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
2WYS
 
 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y, PHOSPHATE ION, ... | | Authors: | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2009-11-20 | | Release date: | 2010-08-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
2JIR
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CYANIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
2JIP
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
2JIO
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
2JIQ
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
