1N2S
 
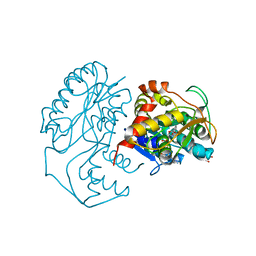 | | CRYSTAL STRUCTURE OF DTDP-6-DEOXY-L-LYXO-4-HEXULOSE REDUCTASE (RMLD) IN COMPLEX WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Blankenfeldt, W, Kerr, I.D, Giraud, M.F, Mcmiken, H.J, Leonard, G.A, Whitfield, C, Messner, P, Graninger, M, Naismith, J.H. | | Deposit date: | 2002-10-24 | | Release date: | 2002-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Variation on a Theme of SDR. dTDP-6-Deoxy-L- lyxo-4-Hexulose Reductase (RmlD) Shows a New Mg(2+)-Dependent Dimerization Mode
Structure, 10, 2002
|
|
7Z1A
 
 | | Nanobody H11 and F2 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2 Nanobody, H11 Nanobody, ... | | Authors: | Mikolajek, H, Naismith, J.H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1C
 
 | | Nanobody H11-B5 and H11-F2 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Nanobody B5, ... | | Authors: | Mikolajek, H, Naismith, J.H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1D
 
 | | Nanobody H11-H6 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, H11-H6 nanobody, ... | | Authors: | Mikolajek, H, Naismith, J.H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1E
 
 | | Nanobody H11-H4 Q98R H100E bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, H11-H4 Q98R H100E, ... | | Authors: | Mikolajek, H, Naismith, J.H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1B
 
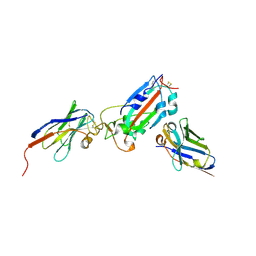 | | Nanobody H11-A10 and F2 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody A10, Nanobody F2, ... | | Authors: | Mikolajek, H, Naismith, J.H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z85
 
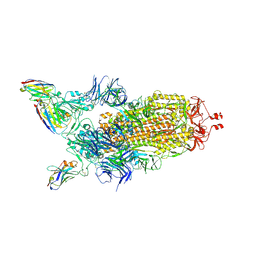 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-B5 nanobody complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-B5, ... | | Authors: | Weckener, M, Naismith, J.H. | | Deposit date: | 2022-03-16 | | Release date: | 2022-07-13 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z9Q
 
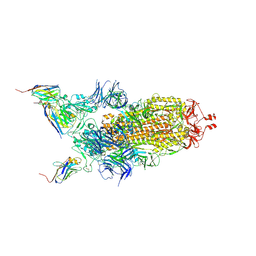 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-A10 nanobody complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-A10, ... | | Authors: | Weckener, M, Naismith, J.H. | | Deposit date: | 2022-03-21 | | Release date: | 2022-07-13 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z7X
 
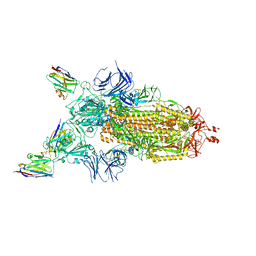 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-H6 nanobody complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-H6, ... | | Authors: | Weckener, M, Naismith, J.H. | | Deposit date: | 2022-03-16 | | Release date: | 2022-07-13 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z86
 
 | |
7Z6V
 
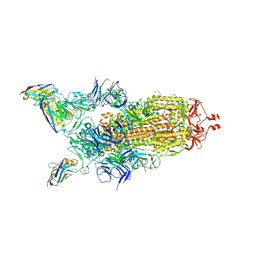 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11 nanobody complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11, ... | | Authors: | Weckener, M, Naismith, J.H, Vogirala, V.K. | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z9R
 
 | |
1V0J
 
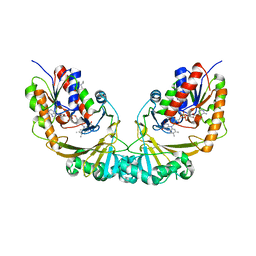 | | Udp-galactopyranose mutase from Mycobacterium tuberculosis | | Descriptor: | BICINE, FLAVIN-ADENINE DINUCLEOTIDE, UDP-GALACTOPYRANOSE MUTASE | | Authors: | Beis, K, Naismith, J.H. | | Deposit date: | 2004-03-30 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UDP-galactopyranose mutase in the oxidised state and Klebsiella pneumoniae UDP-galactopyranose mutase in the (active) reduced state.
J. Mol. Biol., 348, 2005
|
|
6I2J
 
 | | Crystal structure of the ferric enterobactin receptor mutant (Q482A) from Pseudomonas aeruginosa (PfeA) in complex with enterobactin | | Descriptor: | FE (III) ION, Ferric enterobactin receptor, N,N',N''-[(3S,7S,11S)-2,6,10-trioxo-1,5,9-trioxacyclododecane-3,7,11-triyl]tris(2,3-dihydroxybenzamide) | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2018-11-01 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The complex of ferric-enterobactin with its transporter from Pseudomonas aeruginosa suggests a two-site model.
Nat Commun, 10, 2019
|
|
8AP0
 
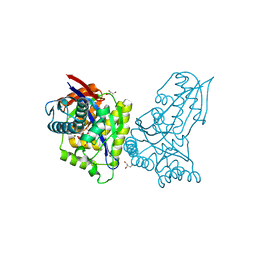 | | ForT Mutant T138V | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 4-azanyl-1~{H}-pyrazole-3,5-dicarboxylic acid, Beta-ribofuranosylaminobenzene 5'-phosphate synthase, ... | | Authors: | Li, W, Naismith, J.H. | | Deposit date: | 2022-08-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental and computational snapshots of C-C bond formation in a C-nucleoside synthase.
Open Biology, 13, 2023
|
|
8AOZ
 
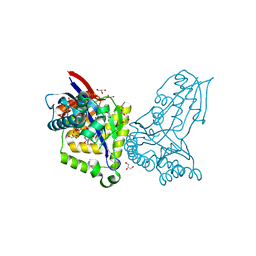 | | ForT Mutant L24A | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-3-[(2~{S},3~{R},4~{S},5~{R})-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]-1~{H}-pyrazole-5-carboxylic acid, Beta-ribofuranosylaminobenzene 5'-phosphate synthase, ... | | Authors: | Li, W, Naismith, J.H. | | Deposit date: | 2022-08-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental and computational snapshots of C-C bond formation in a C-nucleoside synthase.
Open Biology, 13, 2023
|
|
1KD0
 
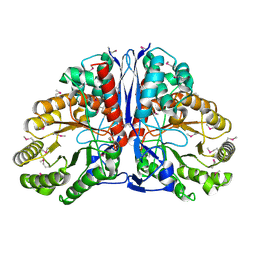 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Apo-structure. | | Descriptor: | 1,2-ETHANEDIOL, beta-methylaspartase | | Authors: | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | Deposit date: | 2001-11-12 | | Release date: | 2001-12-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
1KCZ
 
 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Mg-complex. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, beta-methylaspartase | | Authors: | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | Deposit date: | 2001-11-12 | | Release date: | 2001-12-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
1WA4
 
 | |
6H7V
 
 | |
6HCP
 
 | | Crystal structure of BauA, the Ferric preacinetobactin receptor from Acinetobacter baumannii | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, BauA, ... | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Preacinetobactin not acinetobactin is essential for iron uptake by the BauA transporter of the pathogenAcinetobacter baumannii.
Elife, 7, 2018
|
|
7NHR
 
 | |
6H7F
 
 | | Crystal structure of BauA, the Ferric preacinetobactin receptor from Acinetobacter baumannii in complex with Fe3+-Preacinetobactin-acinetobactin | | Descriptor: | (4~{S},5~{R})-2-[2,3-bis(oxidanyl)phenyl]-~{N}-[2-(1~{H}-imidazol-4-yl)ethyl]-5-methyl-~{N}-oxidanyl-4,5-dihydro-1,3-oxazole-4-carboxamide, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, ... | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Preacinetobactin not acinetobactin is essential for iron uptake by the BauA transporter of the pathogenAcinetobacter baumannii.
Elife, 7, 2018
|
|
5OUT
 
 | |
1WA3
 
 | | Mechanism of the Class I KDPG aldolase | | Descriptor: | 2-KETO-3-DEOXY-6-PHOSPHOGLUCONATE ALDOLASE, PYRUVIC ACID, SULFATE ION | | Authors: | Fullerton, S.W.B, Griffiths, J.S, Merkel, A.B, Wymer, N.J, Hutchins, M.J, Fierke, C.A, Toone, E.J, Naismith, J.H. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of the Class I Kdpg Aldolase.
Bioorg.Med.Chem., 14, 2006
|
|
