4IRC
 
 | | Polymerase-DNA complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Nair, D.T, Sharma, A. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli.
Nucleic Acids Res., 41, 2013
|
|
4IRK
 
 | | structure of Polymerase-DNA complex, dna | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*TP*A*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*(DOC))-3'), DNA (5'-D(*TP*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Nair, D.T, Sharma, A. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli.
Nucleic Acids Res., 41, 2013
|
|
4IRD
 
 | | Structure of Polymerase-DNA complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Nair, D.T, Sharma, A. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli.
Nucleic Acids Res., 41, 2013
|
|
3BJY
 
 | | Catalytic core of Rev1 in complex with DNA (modified template guanine) and incoming nucleotide | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*DAP*DTP*DCP*DCP*DTP*DCP*DCP*DCP*DCP*DTP*DAP*(DOC))-3'), DNA (5'-D(*DTP*DAP*DAP*(P)P*DGP*DTP*DAP*DGP*DGP*DGP*DGP*DAP*DGP*DGP*DAP*DT)-3'), ... | | Authors: | Nair, D.T, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2007-12-05 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Protein-template-directed synthesis across an acrolein-derived DNA adduct by yeast Rev1 DNA polymerase
Structure, 16, 2008
|
|
4DEZ
 
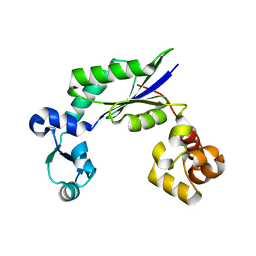 | | Structure of MsDpo4 | | Descriptor: | DNA polymerase IV 1 | | Authors: | Nair, D.T, Sharma, A. | | Deposit date: | 2012-01-22 | | Release date: | 2012-08-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The PAD region in the mycobacterial DinB homologue MsPolIV exhibits positional heterogeneity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4H0E
 
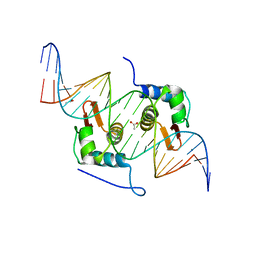 | | Crystal Structure of mutant ORR3 in complex with NTD of AraR | | Descriptor: | 5'-D(*AP*AP*AP*TP*TP*TP*GP*TP*CP*CP*GP*TP*AP*CP*AP*TP*TP*TP*TP*AP*T)-3', 5'-D(*TP*AP*TP*AP*AP*AP*AP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*AP*AP*TP*T)-3', ACETATE ION, ... | | Authors: | Nair, D.T, Jain, D. | | Deposit date: | 2012-09-08 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Spacing between core recognition motifs determines relative orientation of AraR monomers on bipartite operators.
Nucleic Acids Res., 41, 2013
|
|
3OSP
 
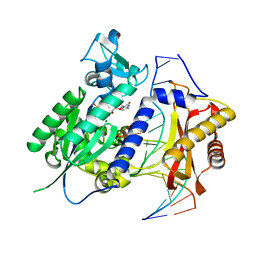 | | Structure of rev1 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*TP*CP*CP*TP*CP*CP*CP*CP*TP*AP*(DOC))-3', 5'-D(*TP*AP*AP*(3DR)P*GP*TP*AP*GP*GP*GP*GP*AP*GP*GP*AP*T)-3', ... | | Authors: | Nair, D.T, Aggarwal, A.K. | | Deposit date: | 2010-09-09 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA synthesis across an abasic lesion by yeast REV1 DNA polymerase.
J.Mol.Biol., 406, 2011
|
|
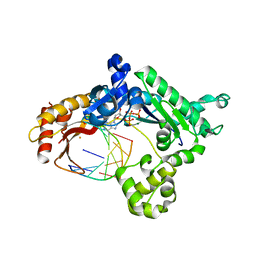
3G6V
 
 | |
3G6Y
 
 | |
3G6X
 
 | |
7WVL
 
 | |
8JRB
 
 | |
6JBP
 
 | | Structure of MP-4 from Mucuna pruriens at 2.22 Angstroms | | Descriptor: | Kunitz-type trypsin inhibitor-like 2 protein | | Authors: | Jain, A, Shikhi, M, Kumar, A, Kumar, A, Nair, D.T, Salunke, D.M. | | Deposit date: | 2019-01-26 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.217 Å) | | Cite: | The structure of MP-4 from Mucuna pruriens at 2.22 angstrom resolution.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
3NCV
 
 | | NgoL | | Descriptor: | DNA mismatch repair protein mutL | | Authors: | Namadurai, S, Jain, D, Nair, D.T. | | Deposit date: | 2010-06-05 | | Release date: | 2010-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The C-terminal domain of the MutL homolog from Neisseria gonorrhoeae forms an inverted homodimer
Plos One, 5, 2010
|
|
4R8U
 
 | | S-SAD structure of DINB-DNA Complex | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA, DNA polymerase IV, ... | | Authors: | Kottur, J, Nair, D.T, Weinert, T, Oligeric, V, Wang, M. | | Deposit date: | 2014-09-03 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination
Nat.Methods, 12, 2015
|
|
5DSS
 
 | |
5D4S
 
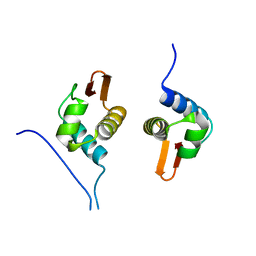 | | Crystal Structure of AraR(DBD) in complex with operator ORX1 | | Descriptor: | Arabinose metabolism transcriptional repressor, DNA (5'-D(*AP*AP*AP*TP*AP*CP*AP*TP*AP*CP*GP*TP*AP*CP*AP*AP*AP*TP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*AP*TP*TP*TP*GP*TP*AP*CP*GP*TP*AP*TP*GP*TP*AP*TP*T)-3') | | Authors: | Jain, D, Narayanan, N, Nair, D.T. | | Deposit date: | 2015-08-08 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Plasticity in Repressor-DNA Interactions Neutralizes Loss of Symmetry in Bipartite Operators.
J.Biol.Chem., 291, 2016
|
|
5D4R
 
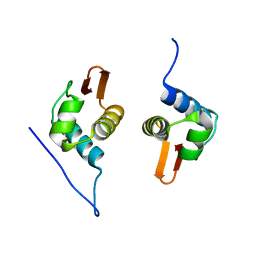 | | Crystal Structure of AraR(DBD) in complex with operator ORE1 | | Descriptor: | Arabinose metabolism transcriptional repressor, DNA (5'-D(*AP*TP*AP*TP*TP*TP*GP*TP*AP*CP*GP*TP*AP*CP*TP*AP*AP*TP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*TP*AP*GP*TP*AP*CP*GP*TP*AP*CP*AP*AP*AP*TP*A)-3') | | Authors: | Jain, D, Narayanan, N, Nair, D.T. | | Deposit date: | 2015-08-08 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Plasticity in Repressor-DNA Interactions Neutralizes Loss of Symmetry in Bipartite Operators.
J.Biol.Chem., 291, 2016
|
|
6IG1
 
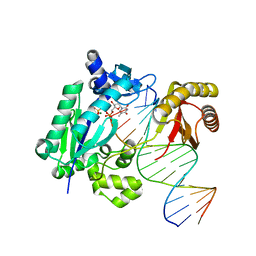 | | DNA polymerase IV - DNA ternary complex 10 | | Descriptor: | DIPHOSPHATE, DNA polymerase IV, DTN3, ... | | Authors: | Kottur, J, Nair, D.T. | | Deposit date: | 2018-09-21 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction.
Nucleic Acids Res., 46, 2018
|
|
6K0W
 
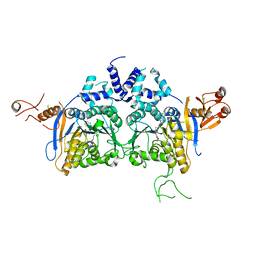 | | DNA methyltransferase in complex with sinefungin | | Descriptor: | Adenine specific DNA methyltransferase (Mod), SINEFUNGIN | | Authors: | Narayanan, N, Nair, D.T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Tetramerization at Low pH Licenses DNA Methylation Activity of M.HpyAXI in the Presence of Acid Stress.
J.Mol.Biol., 432, 2020
|
|
4MTP
 
 | | RdRp from Japanesese Encephalitis Virus | | Descriptor: | RNA dependent RNA polymerase, ZINC ION | | Authors: | Surana, P, Nair, D.T. | | Deposit date: | 2013-09-20 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state.
Nucleic Acids Res., 42, 2014
|
|
4Q43
 
 | | Polymerase-damaged DNA complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*T*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*TP*CP*TP*(RDG)P*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Kottur, J, Sharma, A, Nair, D.T. | | Deposit date: | 2014-04-13 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Unique structural features in DNA polymerase IV enable efficient bypass of the N2 adduct induced by the nitrofurazone antibiotic
Structure, 23, 2015
|
|
4Q45
 
 | | DNA Polymerase- damaged DNA complex | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*TP*CP*TP*A*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*TP*CP*TP*AP*GP*GP*(RDG)P*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Kottur, J, Sharma, A, Nair, D.T. | | Deposit date: | 2014-04-13 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.176 Å) | | Cite: | Unique structural features in DNA polymerase IV enable efficient bypass of the N2 adduct induced by the nitrofurazone antibiotic
Structure, 23, 2015
|
|
5YK4
 
 | | Mismatch Repair Protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS | | Authors: | Nirwal, S, Nair, D.T. | | Deposit date: | 2017-10-12 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Mechanism of formation of a toroid around DNA by the mismatch sensor protein.
Nucleic Acids Res., 46, 2018
|
|
5YJS
 
 | | Structure of vicilin from Capsicum annuum | | Descriptor: | 2-HYDROXYBENZOIC ACID, CHLORIDE ION, COPPER (I) ION, ... | | Authors: | Shikhi, M, Nair, D.T, Salunke, D.M. | | Deposit date: | 2017-10-11 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-guided identification of function: role ofCapsicum annuumvicilin during oxidative stress.
Biochem. J., 475, 2018
|
|
