6TWZ
 
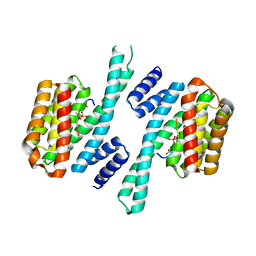 | | 14-3-3 sigma complexed with a phosphorylated 16E6 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D(-)-TARTARIC ACID, ... | | Authors: | Gogl, G, Cousido-Siah, A, Sluchanko, N.N, Trave, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual Specificity PDZ- and 14-3-3-Binding Motifs: A Structural and Interactomics Study.
Structure, 28, 2020
|
|
2QC7
 
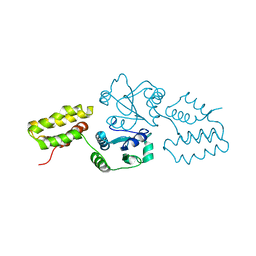 | | Crystal structure of the protein-disulfide isomerase related chaperone ERp29 | | Descriptor: | Endoplasmic reticulum protein ERp29 | | Authors: | Barak, N.N, Sevvana, M, Neumann, P, Malesevic, M, Naumann, K, Fischer, G, Sheldrick, G.M, Stubbs, M.T, Ferrari, D.M. | | Deposit date: | 2007-06-19 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and functional analysis of the protein disulfide isomerase-related protein ERp29.
J.Mol.Biol., 385, 2009
|
|
5JH9
 
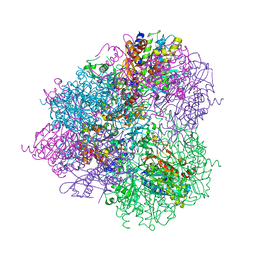 | | Crystal structure of prApe1 | | Descriptor: | CACODYLATE ION, Vacuolar aminopeptidase 1, ZINC ION | | Authors: | Noda, N.N, Adachi, W, Inagaki, F. | | Deposit date: | 2016-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
8QPT
 
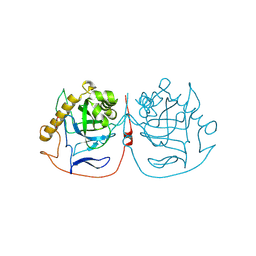 | | Crystal structure of pyrophosphatase from Ogataea parapolymorpha | | Descriptor: | GLYCEROL, MAGNESIUM ION, inorganic diphosphatase | | Authors: | Matyuta, I.O, Rodina, E.V, Vorobyeva, N.N, Kurilova, S.A, Bezpalaya, E.Y, Boyko, K.M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of yeast mitochondrial type pyrophosphatase provides a model to study pathological mutations in its human ortholog.
Biochem.Biophys.Res.Commun., 738, 2024
|
|
3L2B
 
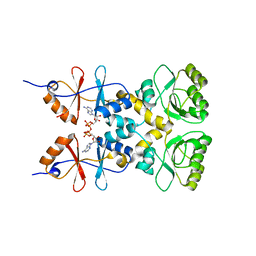 | | Crystal structure of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with activator, diadenosine tetraphosphate | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Probable manganase-dependent inorganic pyrophosphatase | | Authors: | Tuominen, H, Salminen, A, Oksanen, E, Jamsen, J, Heikkila, O, Lehtio, L, Magretova, N.N, Goldman, A, Baykov, A.A, Lahti, R. | | Deposit date: | 2009-12-15 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridiumperfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate.
J.Mol.Biol., 2010
|
|
1HQ5
 
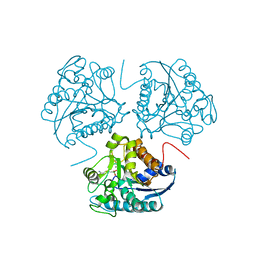 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH S-(2-BORONOETHYL)-L-CYSTEINE, AN L-ARGININE ANALOGUE | | Descriptor: | ARGINASE 1, MANGANESE (II) ION, S-2-(BORONOETHYL)-L-CYSTEINE | | Authors: | Kim, N.N, Cox, J.D, Baggio, R.F, Emig, F.A, Mistry, S.K, Harper, S.L, Speicher, D.W, Morris Jr, S.M, Ash, D.E, Traish, A, Christianson, D.W. | | Deposit date: | 2000-12-14 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing erectile function: S-(2-boronoethyl)-L-cysteine binds to arginase as a transition state analogue and enhances smooth muscle relaxation in human penile corpus cavernosum.
Biochemistry, 40, 2001
|
|
4N9O
 
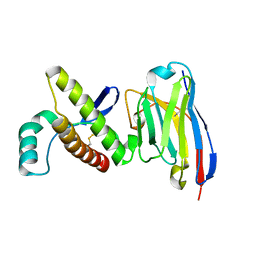 | | Probing the N-terminal beta-sheet conversion in the crystal structure of the human prion protein bound to a Nanobody | | Descriptor: | Major prion protein, Nanobody Nb484 | | Authors: | Abskharon, R.N.N, Giachin, G, Wohlkonig, A, Soror, S.H, Pardon, E, Legname, G, Steyaert, J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the N-Terminal beta-Sheet Conversion in the Crystal Structure of the Human Prion Protein Bound to a Nanobody.
J.Am.Chem.Soc., 136, 2014
|
|
4A56
 
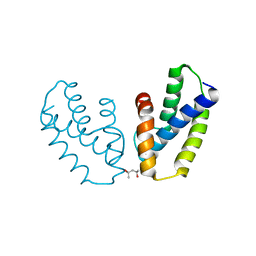 | | Crystal structure of the type 2 secretion system pilotin from Klebsiella Oxytoca | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PULLULANASE SECRETION PROTEIN PULS | | Authors: | Tosi, T, Nickerson, N.N, Mollica, L, RingkjobingJensen, M, Blackledge, M, Baron, B, England, P, Pugsley, A.P, Dessen, A. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Pilotin-Secretin Recognition in the Type II Secretion System of Klebsiella Oxytoca.
Mol.Microbiol, 82, 2011
|
|
1E52
 
 | | Solution structure of Escherichia coli UvrB C-terminal domain | | Descriptor: | EXCINUCLEASE ABC SUBUNIT | | Authors: | Alexandrovich, A.A, Kelly, G.G, Frenkiel, T.A, Moolenaar, G.F, Goosen, N.N, Sanderson, M.R, Lane, A.N. | | Deposit date: | 2000-07-14 | | Release date: | 2001-07-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Hydrodynamics and Thermodynamics of the Uvrb C-Terminal Domain.
J.Biomol.Struct.Dyn., 19, 2001
|
|
2AHE
 
 | | Crystal structure of a soluble form of CLIC4. intercellular chloride ion channel | | Descriptor: | Chloride intracellular channel protein 4 | | Authors: | Littler, D.R, Assaad, N.N, Harrop, S.J, Brown, L.J, Pankhurst, G.J, Luciani, P, Aguilar, M.-I, Mazzanti, M, Berryman, M.A, Breit, S.N, Curmi, P.M.G. | | Deposit date: | 2005-07-28 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the soluble form of the redox-regulated chloride ion channel protein CLIC4.
FEBS J., 272, 2005
|
|
6XZT
 
 | | Crystal structure of helicase Pif1 from Thermus oshimai mutant G110C-E410C | | Descriptor: | PIF1 helicase | | Authors: | Dai, Y.X, Chen, W.F, Teng, F.Y, Liu, N.N, Hou, X.M, Dou, S.X, Rety, S, Xi, X.G. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.341 Å) | | Cite: | Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition.
Nucleic Acids Res., 49, 2021
|
|
7OAR
 
 | | Crystal structure of helicase Pif1 from Thermus oshimai in complex with parallel G-quadruplex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (28-MER), MAGNESIUM ION, ... | | Authors: | Dai, Y.X, Liu, N.N, Guo, H.L, Chen, W.F, Rety, S, Xi, X.G. | | Deposit date: | 2021-04-20 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural mechanism underpinning Thermus oshimai Pif1-mediated G-quadruplex unfolding.
Embo Rep., 23, 2022
|
|
8C18
 
 | | Solution structure of carotenoid-binding protein AstaPo1 in complex with astaxanthin | | Descriptor: | ASTAXANTHIN, Astaxanthin binding fasciclin family protein | | Authors: | Kornilov, F.D, Savitskaya, A.G, Slonimskiy, Y.B, Goncharuk, S.A, Sluchanko, N.N, Mineev, K.S. | | Deposit date: | 2022-12-20 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the ligand promiscuity of the neofunctionalized, carotenoid-binding fasciclin domain protein AstaP.
Commun Biol, 6, 2023
|
|
5W2F
 
 | | Crystal Structure of the C-terminal Domain of Human eIF2D at 1.4 A resolution | | Descriptor: | Eukaryotic translation initiation factor 2D, FORMIC ACID | | Authors: | Vaidya, A.T, Lomakin, I.B, Joseph, N.N, Dmitriev, S.E, Steitz, T.A. | | Deposit date: | 2017-06-06 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the C-terminal Domain of Human eIF2D and Its Implications on Eukaryotic Translation Initiation.
J. Mol. Biol., 429, 2017
|
|
6ZFD
 
 | | 14-3-3 zeta bound to the phosphorylated 18E6 C-terminus | | Descriptor: | 14-3-3 protein zeta/delta,Protein E6, GLYCEROL | | Authors: | Gogl, G, Tugaeva, K.V, Sluchanko, N.N, Trave, G. | | Deposit date: | 2020-06-17 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hierarchized phosphotarget binding by the seven human 14-3-3 isoforms.
Nat Commun, 12, 2021
|
|
6ZFG
 
 | | 14-3-3 zeta chimera with 18E6 and fusicoccin | | Descriptor: | 14-3-3 protein zeta/delta,Protein E6, FUSICOCCIN, GLYCEROL | | Authors: | Gogl, G, Tugaeva, K, Sluchanko, N.N, Trave, G. | | Deposit date: | 2020-06-17 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Hierarchized phosphotarget binding by the seven human 14-3-3 isoforms.
Nat Commun, 12, 2021
|
|
3L31
 
 | | Crystal structure of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with the inhibitor, AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Probable manganase-dependent inorganic pyrophosphatase | | Authors: | Tuominen, H, Salminen, A, Oksanen, E, Jamsen, J, Heikkila, O, Lehtio, L, Magretova, N.N, Goldman, A, Baykov, A.A, Lahti, R. | | Deposit date: | 2009-12-16 | | Release date: | 2010-04-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridiumperfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate.
J.Mol.Biol., 2010
|
|
8ZES
 
 | | Crystal structure of the Wuhan SARS-CoV-2 RBD (333-541) complexed with P2C5 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody P2C5, ... | | Authors: | Sluchanko, N.N, Varfolomeeva, L.A, Shcheblyakov, D.V, Logunov, D.Y, Gintsburg, A.L, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-05-06 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis for Evasion of New SARS-CoV-2 Variants from the Potent Virus-Neutralizing Nanobody Targeting the S-Protein Receptor-Binding Domain.
Biochemistry Mosc., 89, 2024
|
|
8JYE
 
 | | Crystal Structure of Intracellular B30.2 Domain of BTN3A1 and BTN2A1 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, Butyrophilin subfamily 2 member A1, ... | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.-C, Guo, R.-T, Zhang, Y.H. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8ZER
 
 | | Crystal structure of the complex of Wuhan SARS-CoV-2 RBD (319-541) with P2C5 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody P2C5, Spike protein S1, ... | | Authors: | Sluchanko, N.N, Varfolomeeva, L.A, Shcheblyakov, D.V, Logunov, D.Y, Gintsburg, A.L, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-05-06 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Evasion of New SARS-CoV-2 Variants from the Potent Virus-Neutralizing Nanobody Targeting the S-Protein Receptor-Binding Domain.
Biochemistry Mosc., 89, 2024
|
|
8JYC
 
 | | Crystal Structure of Intracellular B30.2 Domain of BTN3A1 and BTN2A1 in Complex with DMAPP | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 2 member A1, Butyrophilin subfamily 3 member A1, ... | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.-C, Guo, R.-T, Zhang, Y.H. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
7XUP
 
 | | Crystal structure of TPe3.0 | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Chen, X, Qian, J.Y, Sun, N.N, Zhong, F.R, Wu, Y.Z. | | Deposit date: | 2022-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Enantioselective [2+2]-cycloadditions with triplet photoenzymes.
Nature, 611, 2022
|
|
7XUQ
 
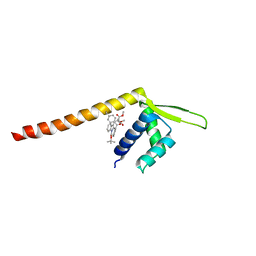 | | Crystal structure of Tpe3.0 complexed with N-Boc-3-alkenylindole | | Descriptor: | Transcriptional regulator, PadR-like family, dimethyl 2-[[2-methyl-1-[(2-methylpropan-2-yl)oxycarbonyl]indol-3-yl]methyl]-2-prop-2-enyl-propanedioate | | Authors: | Chen, X, Qian, J.Y, Sun, N.N, Zhong, F.R, Wu, Y.Z. | | Deposit date: | 2022-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enantioselective [2+2]-cycloadditions with triplet photoenzymes.
Nature, 611, 2022
|
|
5H9W
 
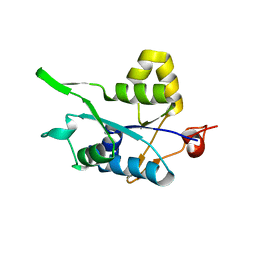 | | Crystal structure of Regnase PIN domain, form II | | Descriptor: | Ribonuclease ZC3H12A, SODIUM ION | | Authors: | Yokogawa, M, Tsushima, T, Adachi, W, Noda, N.N, Inagaki, F. | | Deposit date: | 2015-12-29 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
4J8R
 
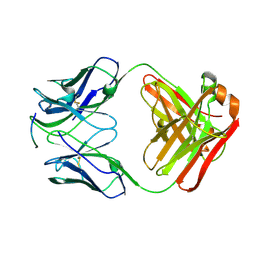 | | Structure of an octapeptide repeat of the prion protein bound to the POM2 Fab antibody fragment | | Descriptor: | Heavy chain of POM2 Fab, Light chain of POM2 Fab, Major prion protein | | Authors: | Swayampakula, M, Baral, P.K, Kav, N.N.V, Aguzzi, A, James, M.N.G. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | The crystal structure of an octapeptide repeat of the Prion protein in complex with a Fab fragment of the POM2 antibody.
Protein Sci., 22, 2013
|
|
