5J7K
 
 | |
5J54
 
 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | Carotenoid oxygenase, FE (III) ION, OXYGEN MOLECULE, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
7QRD
 
 | | Crystal structure of mouse CARM1 in complex with histone H3_10-25 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-01-11 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CARM1 Transition State Mimics
To Be Published
|
|
5JDU
 
 | | Crystal structure for human thrombin mutant D189A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pozzi, N, Chen, Z, Di Cera, E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Loop Electrostatics Asymmetry Modulates the Preexisting Conformational Equilibrium in Thrombin.
Biochemistry, 55, 2016
|
|
3BLL
 
 | | TGT mutant in complex with Boc-preQ1 | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION, ... | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2007-12-11 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor
Plos One, 8, 2013
|
|
7QPH
 
 | | Crystal structure of mouse CARM1 in complex with histone H3_22-31 K27 acetylated | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-ETHANEDIOL, Histone H3 22-31 K27 acetylated, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-01-04 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CARM1 Transition State Mimics
To Be Published
|
|
3BLO
 
 | | TGT mutant in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2007-12-11 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor
Plos One, 8, 2013
|
|
5IYE
 
 | | Comparison of X-ray crystal structures of a tetradecamer sequence d(CCCGGGTACCCGGG)2 at 1.7 resolution | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*GP*TP*AP*CP*CP*CP*GP*GP*G)-3'), ZINC ION | | Authors: | Karthik, S, Thirugnanasambandam, A, Mandal, P.K, Gautham, N. | | Deposit date: | 2016-03-24 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Comparison of X-ray crystal structures of a tetradecamer sequence d(CCCGGGTACCCGGG)2 at 1.7 angstrom resolution.
Nucleosides Nucleotides Nucleic Acids, 36, 2017
|
|
5IYG
 
 | |
5IYJ
 
 | |
3ZNU
 
 | | Crystal structure of ClcF in crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION, ... | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
3BLD
 
 | | tRNA guanine transglycosylase V233G mutant preQ1 complex structure | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2007-12-11 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor
Plos One, 8, 2013
|
|
3BWJ
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor lead compound Arc-1034 | | Descriptor: | (2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-N-(6-{[(1R)-4-carbamimidamido-1-{[(1R)-4-carbamimidamido-1-carbamoylbutyl]carbamoyl}butyl]amino}-6-oxohexyl)-3,4-dihydroxytetrahydrofuran-2-carboxamide, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Lavogina, D, Koenig, N, Uri, A, Bossemeyer, D. | | Deposit date: | 2008-01-09 | | Release date: | 2009-02-03 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of ARC-type inhibitor (ARC-1034) binding to protein kinase A catalytic subunit and rational design of bisubstrate analogue inhibitors of basophilic protein kinases.
J.Med.Chem., 52, 2009
|
|
5J0Z
 
 | | Crystal structure of GLIC in complex with DHA | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Basak, S, Schmandt, N, Chakrapani, S. | | Deposit date: | 2016-03-28 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure and dynamics of a lipid-induced potential desensitized-state of a pentameric ligand-gated channel.
Elife, 6, 2017
|
|
5J1W
 
 | | Crystal structure of human CLK1 in complex with pyrido[3,4-g]quinazoline derivative ZW31 (compound 14) | | Descriptor: | Dual specificity protein kinase CLK1, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Esvan, Y.J, Zeinyeh, W, Boibessot, T, Nauton, L, Thery, V, Loaec, N, Meijer, L, Giraud, F, Moreau, P, Anizon, F, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery of pyrido[3,4-g]quinazoline derivatives as CMGC family protein kinase inhibitors: Design, synthesis, inhibitory potency and X-ray co-crystal structure.
Eur.J.Med.Chem., 118, 2016
|
|
5JCP
 
 | | RhoGAP domain of ARAP3 in complex with RhoA in the transition state | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3,Linker,Transforming protein RhoA, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bao, H, Li, F, Wang, C, Wang, N, Jiang, Y, Tang, Y, Wu, J, Shi, Y. | | Deposit date: | 2016-04-15 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Specific Recognition of RhoA by the Dual GTPase-activating Protein ARAP3
J.Biol.Chem., 291, 2016
|
|
7R6X
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2E12 Fab, S309 Fab, and S304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Monoclonal antibody S2E12 Fab heavy chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
3ZNJ
 
 | | Crystal structure of unliganded ClcF from R.opacus 1CP in crystal form 1. | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
5J55
 
 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, Carotenoid oxygenase, FE (III) ION, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5JK5
 
 | |
5JKI
 
 | | Crystal structure of the first transmembrane PAP2 type phosphatidylglycerolphosphate phosphatase from Bacillus subtilis | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative lipid phosphate phosphatase YodM, TUNGSTATE(VI)ION, ... | | Authors: | El Ghachi, M, Howe, N, Lampion, A, Delbrassine, F, Vogeley, L, Caffrey, M, Sauvage, E, Auger, R, Guiseppe, A, Roure, S, Perlier, S, Mengin-lecreulx, D, Foglino, M, Touze, T. | | Deposit date: | 2016-04-26 | | Release date: | 2017-02-22 | | Last modified: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure and biochemical characterization of the transmembrane PAP2 type phosphatidylglycerol phosphate phosphatase from Bacillus subtilis.
Cell. Mol. Life Sci., 74, 2017
|
|
8IZH
 
 | | Crystal structure of trypsin-aminoguanidine complex at 2.30 Angstroms resolution | | Descriptor: | AMINOGUANIDINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ahmad, M.S, Kalam, N, Akbar, Z, Rasheed, S, Choudhary, M.I. | | Deposit date: | 2023-04-07 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the binding of famotidine, cimetidine, guanidine, and pimagedine with serine protease.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
8IZI
 
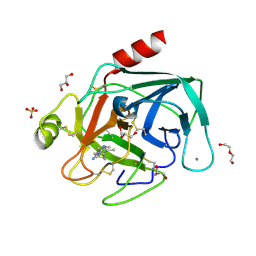 | | Crystal structure of trypsin-cimetidine complex at 2.10 Angstroms resolution | | Descriptor: | 2-cyano-1-methyl-3-[2-[(5-methyl-1~{H}-imidazol-4-yl)methylsulfanyl]ethyl]guanidine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ahmad, M.S, Kalam, N, Akbar, Z, Rasheed, S, Choudhary, M.I. | | Deposit date: | 2023-04-07 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the binding of famotidine, cimetidine, guanidine, and pimagedine with serine protease.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
8IYV
 
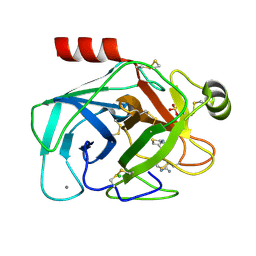 | | Crystal structure of trypsin-famotidine complex at 2.10 Angstroms resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cationic trypsin, ... | | Authors: | Ahmad, M.S, Kalam, N, Akbar, Z, Rasheed, S, Choudhary, M.I. | | Deposit date: | 2023-04-06 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the binding of famotidine, cimetidine, guanidine, and pimagedine with serine protease.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
8IZK
 
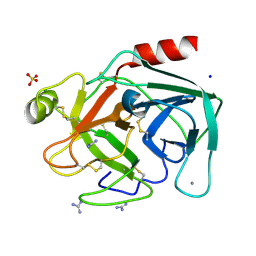 | | Crystal structure of trypsin-guanidine complex at 2.05 Angstroms resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cationic trypsin, ... | | Authors: | Ahmad, M.S, Kalam, N, Akbar, Z, Rasheed, S, Choudhary, M.I. | | Deposit date: | 2023-04-07 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the binding of famotidine, cimetidine, guanidine, and pimagedine with serine protease.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
