4UBA
 
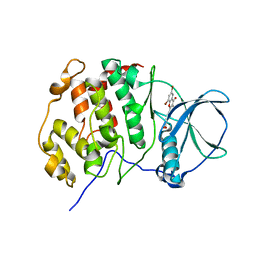 | | Low-salt structure of protein kinase CK2 catalytic subunit with 4'-carboxy-6,8-bromo-flavonol (FLC26) | | Descriptor: | 4-(6,8-dibromo-3-hydroxy-4-oxo-4H-chromen-2-yl)benzoic acid, Casein kinase II subunit alpha | | Authors: | Niefind, K, Bischoff, N, Guerra, B, Golub, A, Issinger, O.-G. | | Deposit date: | 2014-08-12 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | A Note of Caution on the Role of Halogen Bonds for Protein Kinase/Inhibitor Recognition Suggested by High- And Low-Salt CK2 alpha Complex Structures.
Acs Chem.Biol., 10, 2015
|
|
4U4O
 
 | | Crystal structure of Geneticin bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U53
 
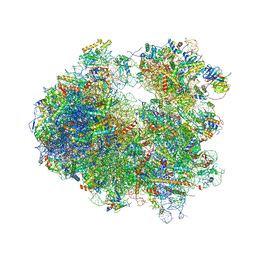 | | Crystal structure of Deoxynivalenol bound to the yeast 80S ribosome | | Descriptor: | (3beta,7alpha)-3,7,15-trihydroxy-12,13-epoxytrichothec-9-en-8-one, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
2VR9
 
 | |
8OSL
 
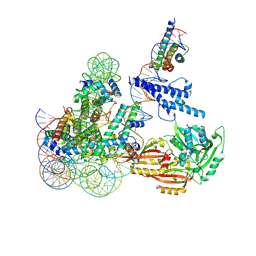 | | Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (147-MER), ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
1LVG
 
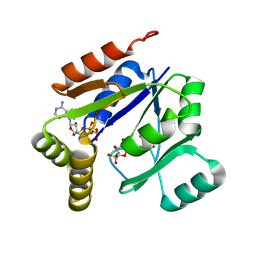 | | Crystal structure of mouse guanylate kinase in complex with GMP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, Guanylate kinase, ... | | Authors: | Sekulic, N, Shuvalova, L, Spangenberg, O, Konrad, M, Lavie, A. | | Deposit date: | 2002-05-28 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the closed
conformation of mouse guanylate kinase.
J.Biol.Chem., 277, 2002
|
|
8OTS
 
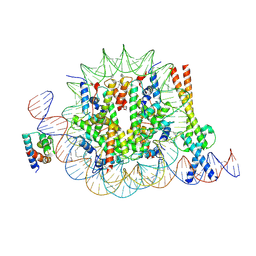 | | OCT4 and MYC-MAX co-bound to a nucleosome | | Descriptor: | DNA (127-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
4U4N
 
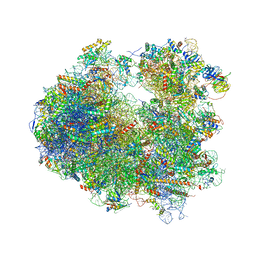 | | Crystal structure of Edeine bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
8OTT
 
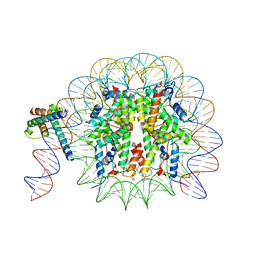 | | MYC-MAX bound to a nucleosome at SHL+5.8 | | Descriptor: | DNA (144-MER), Histone H2A type 1-B/E, Histone H2A type 1-K, ... | | Authors: | Stoos, L, Michael, A.K, Kempf, G, Kater, L, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
2WE0
 
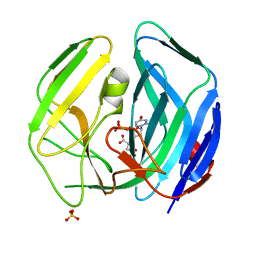 | | EBV dUTPase mutant Cys4Ser | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, MALATE LIKE INTERMEDIATE, ... | | Authors: | Freeman, L, Buisson, M, Tarbouriech, N, Burmeister, W.P. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Flexible Motif V of Epstein-Barr Virus Deoxyuridine 5'-Triphosphate Pyrophosphatase is Essential for Catalysis.
J.Biol.Chem., 284, 2009
|
|
1PGR
 
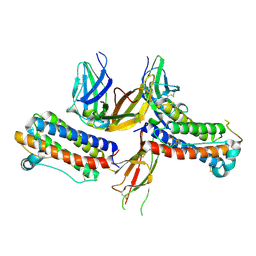 | | 2:2 COMPLEX OF G-CSF WITH ITS RECEPTOR | | Descriptor: | PROTEIN (G-CSF RECEPTOR), PROTEIN (GRANULOCYTE COLONY-STIMULATING FACTOR) | | Authors: | Aritomi, M, Kunishima, N, Okamoto, T, Kuroki, R, Ota, Y, Morikawa, K. | | Deposit date: | 1999-03-08 | | Release date: | 2000-03-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Atomic structure of the GCSF-receptor complex showing a new cytokine-receptor recognition scheme.
Nature, 401, 1999
|
|
4U1F
 
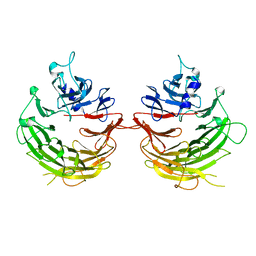 | |
8P7K
 
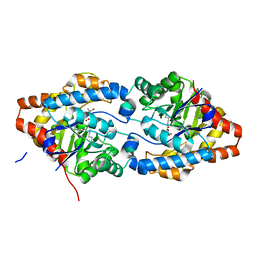 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | (2~{R})-2-methylpentanedioic acid, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Dym, O, Aggawal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-30 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
4U4Q
 
 | | Crystal structure of Homoharringtonine bound to the yeast 80S ribosome | | Descriptor: | (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
1PIU
 
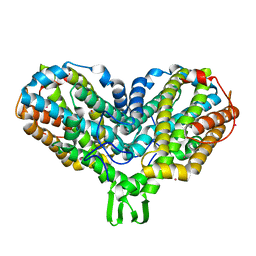 | | OXIDIZED RIBONUCLEOTIDE REDUCTASE R2-D84E MUTANT CONTAINING OXO-BRIDGED DIFERRIC CLUSTER | | Descriptor: | FE (III) ION, MERCURY (II) ION, OXYGEN ATOM, ... | | Authors: | Voegtli, W.C, Khidekel, N, Baldwin, J, Ley, B.A, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Ribonucleotide Reductase R2 Mutant that Accumulates a u-1,2-Peroxodiiron(III)
Intermediate during Oxygen Activation
J.Am.Chem.Soc., 122, 2000
|
|
5NS8
 
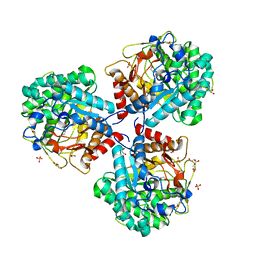 | | Crystal structure of beta-glucosidase BglM-G1 mutant H75R from marine metagenome in complex with inhibitor 1-Deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, GLYCEROL, SULFATE ION, ... | | Authors: | Mhaindarkar, D.C, Gasper, R, Lupilova, N, Leichert, L.I, Hofmann, E. | | Deposit date: | 2017-04-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Loss of a conserved salt bridge in bacterial glycosyl hydrolase BgIM-G1 improves substrate binding in temperate environments.
Commun Biol, 1, 2018
|
|
1PL9
 
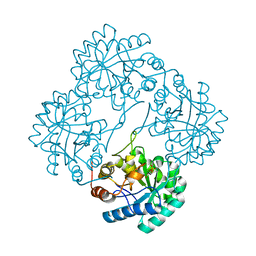 | | Crystal structure of KDO8P synthase in its binary complex with substrate analog Z-FPEP | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, 3-FLUORO-2-(PHOSPHONOOXY)PROPANOIC ACID | | Authors: | Vainer, R, Adir, N, Baasov, T, Belakhov, V, Rabkin, E. | | Deposit date: | 2003-06-08 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of KDO8P synthase in its binary complex with substrate analog Z-FPEP
To be Published
|
|
2VZ9
 
 | |
4U4R
 
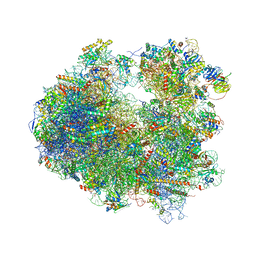 | | Crystal structure of Lactimidomycin bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R,5S,6E)-2-hydroxy-5-methyl-7-[(2R,3S,4E,6Z,10E)-3-methyl-12-oxooxacyclododeca-4,6,10-trien-2-yl]-4-oxooct-6-en-1-yl}piperidine-2,6-dione, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U94
 
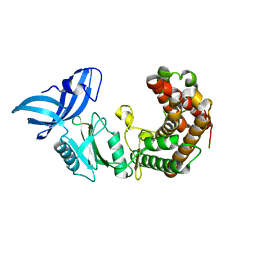 | | Structure of mycobacterial maltokinase, the missing link in the essential GlgE-pathway | | Descriptor: | MAGNESIUM ION, Maltokinase | | Authors: | Fraga, J, Empadinhas, N, Pereira, P.J.B, Macedo-Ribeiro, S. | | Deposit date: | 2014-08-05 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.473 Å) | | Cite: | Structure of mycobacterial maltokinase, the missing link in the essential GlgE-pathway.
Sci Rep, 5, 2015
|
|
4UFC
 
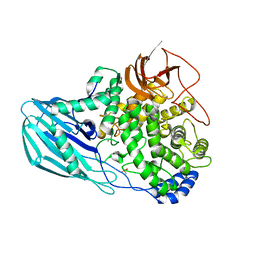 | | Crystal structure of the GH95 enzyme BACOVA_03438 | | Descriptor: | CACODYLATE ION, CALCIUM ION, GH95, ... | | Authors: | Rogowski, A, Briggs, J.A, Mortimer, J.C, Tryfona, T, Terrapon, N, Lowe, E.C, Basle, A, Morland, C, Day, A.M, Zheng, H, Rogers, T.E, Thompson, P, Hawkins, A.R, Yadav, M.P, Henrissat, B, Martens, E.C, Dupree, P, Gilbert, H.J, Bolam, D.N. | | Deposit date: | 2015-03-16 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Glycan Complexity Dictates Microbial Resource Allocation in the Large Intestine.
Nat.Commun., 6, 2015
|
|
4W2I
 
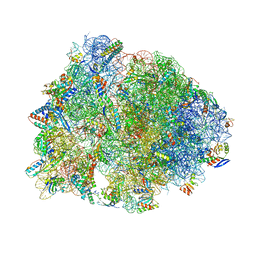 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with negamycin, mRNA and three deacylated tRNAs in the A, P and E sites | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Szal, T, Jiang, F, Gupta, P, Matsuda, R, Shiozuka, M, Steitz, T.A, Vazquez-Laslop, N, Mankin, A.S. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Negamycin Interferes with Decoding and Translocation by Simultaneous Interaction with rRNA and tRNA.
Mol.Cell, 56, 2014
|
|
4V8C
 
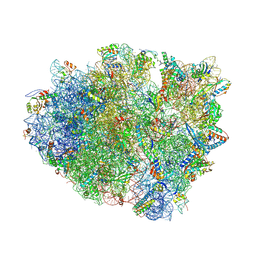 | | Crystal structure analysis of ribosomal decoding (near-cognate tRNA-leu complex with paromomycin). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
1MQB
 
 | | Crystal Structure of Ephrin A2 (ephA2) Receptor Protein Kinase | | Descriptor: | Ephrin type-A receptor 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Nowakowski, J, Cronin, C.N, McRee, D.E, Knuth, M.W, Nelson, C, Pavletich, N, Rogers, J, Sang, B.C, Scheibe, D.N, Swanson, R.V, Thompson, D.A. | | Deposit date: | 2002-09-16 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Cancer Related Aurora-A, FAK and EphA2 Protein Kinases from Nanovolume Crystallography
Structure, 10, 2003
|
|
4V26
 
 | | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context- dependent cytostasis in cancer cells | | Descriptor: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, N-[4-(2-CHLORO-5-METHYLPYRIMIDIN-4-YL)PHENYL]-2,4-DIHYDROXY-N-(4-{[(TRIFLUOROACETYL)AMINO]METHYL}BENZYL)BENZAMIDE, ... | | Authors: | Moore, J.D, Staniszewska, A, Shaw, T, D'Alessandro, J, Davis, B, Surgenor, A, Baker, L, Matassova, N, Murray, J, Macias, A, Brough, P, Wood, M, Mahon, P.C. | | Deposit date: | 2014-10-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context-dependent cytostasis in cancer cells.
Oncotarget, 5, 2014
|
|
